Last Updated: March 7, 2024
Introduction to Lipid Synthesis Pathways
All cells possess the capacity to synthesize fatty acids and phospholipids as a means to ensure continuous integrity of existing membranes as well as in the synthesis of new membranes. However, the majority of fatty acid biosynthesis occurs in hepatocytes and adipocytes.
Synthesis of triglycerides occurs in most cells but predominantly occurs in intestinal enterocytes for the delivery of dietary fatty acids to the body and hepatocytes of the liver for the delivery of endogenous fatty acids to, primarily, cardiac and skeletal muscle and to adipocytes. Adipocytes can be considered the major cell types tasked with triglyceride synthesis as these are the primary fatty acid storage cells the body.
Fatty acid, triglyceride, and phospholipid synthesis represent reductive biosynthetic processes and as such utilize NADPH as the co-factor for the reductive reactions. The major sources of the NADPH are the dehydrogenases of the Pentose Phosphate Pathway and the the malic enzyme catalyzed oxidation of cytoplasmic malate to pyruvate as outlined in the Figure in the next section and discussed in detail below.
The synthesis of fatty acids takes place in the cytosol. The synthesis of triglycerides takes place within the endoplasmic reticulum, ER. The synthesis of phospholipids occurs on the cytoplasmic face of the membranes of the ER.
Origin of Cytoplasmic Acetyl-CoA
The cytoplasmic acetyl-CoA that is required for fatty acid biosynthesis (and cholesterol biosynthesis) is initially generated in the mitochondria primarily from two sources, the pyruvate dehydrogenase (PDH) reaction and fatty acid oxidation, but is also derived from mitochondrial amino acid catabolism. In order for these acetyl-CoA units to be utilized for fatty acid synthesis they must be present in the cytoplasm. The shift from fatty acid oxidation and glycolytic oxidation, as well as amino acid metabolism, occurs when the need for energy diminishes. This results in reduced oxidation of acetyl-CoA in the TCA cycle and the re-oxidation of reduced NADH and FADH2 via the oxidative phosphorylation pathway. Under these conditions the mitochondrial 2-carbon acetyl units can be stored as fat for future energy demands or diverted into cholesterol biosynthesis.
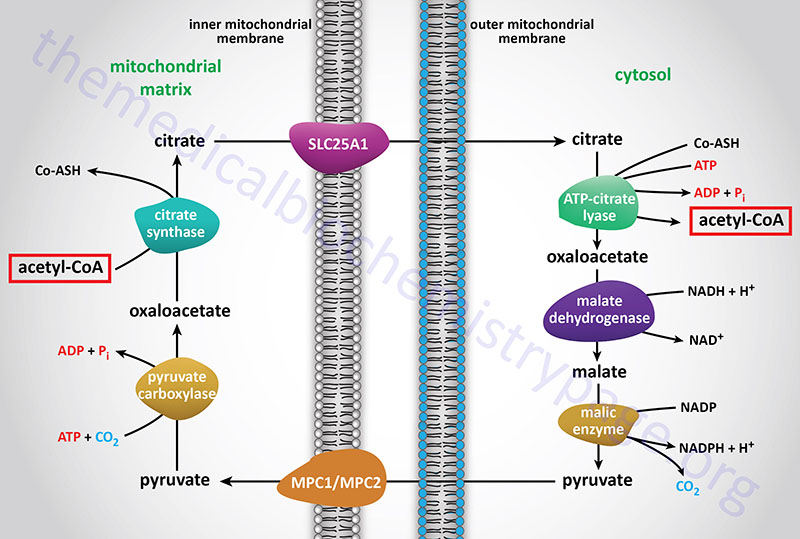
Acetyl-CoA enters the cytoplasm in the form of citrate via the tricarboxylate transport system (encoded by the SLC25A1 gene; see Figure above). In the cytoplasm, citrate and coenzyme A (CoASH) are converted to oxaloacetate and acetyl-CoA by the ATP driven ATP-citrate lyase reaction. This reaction is essentially the reverse of that catalyzed by the TCA cycle enzyme, citrate synthase, except it requires the energy of ATP hydrolysis to drive it forward. The resultant oxaloacetate is converted to malate by cytoplasmic malate dehydrogenase (encoded by the MDH1 gene).
ATP-citrate lyase is encoded by the ACLY gene which is located on chromosome 17q21.2 and is composed of 30 exons that generate four alternatively spliced mRNAs, each of which encodes a unique protein isoform.
Functional ATP-citrate lyase is a homotetrameric complex. ATP-citrate lyase is a critical metabolic enzyme that links glucose metabolism to the processes of both fatty acid synthesis and cholesterol synthesis. In addition to the role of ATP-citrate lyase in cytosolic lipid synthesis, the enzyme is found in the nucleus where it plays a role in epigenetic control of gene expression by providing the acetyl-CoA used in histone acetylation.
The malate produced by this pathway can undergo oxidative decarboxylation by cytoplasmic malic enzyme (encoded by the ME1 gene). The co-enzyme for this reaction is NADP+ generating NADPH. The advantage of this series of reactions for converting mitochondrial acetyl-CoA into cytoplasmic acetyl-CoA is that the NADPH produced by the cytosolic malic enzyme reaction can be a major source of reducing co-factor for the enzymatic activities of the fatty acid synthase (FAS) complex.
Malic Enzymes
Humans express three malic enzymes, one cytoplasmic that requires NADP+ and two mitochondrial enzymes, one that requires NADP+ and one that requires NAD+.
The NADP+-dependent cytoplasmic enzyme is called malic enzyme 1 and is encoded by the ME1 gene. The ME1 gene is located on chromosome 6q14.2 and is composed of 14 exons that encode a protein of 572 amino acids. The highest level of ME1 expression is in adipose tissue attesting to the major role of the ME1 enzyme in fatty acid synthesis. The NADPH produced by the ME1 catalyzed reaction can be used in biomass producing reductive biosynthetic reactions such as fatty synthesis, cholesterol synthesis, and phospholipid synthesis.
The NAD+-dependent mitochondrial enzyme is called malic enzyme 2 and is encoded by the ME2 gene. The ME2 gene is located on chromosome 18q21.2 and is composed of 16 exons that generate two alternatively spliced mRNAs encoding precursor proteins of 584 amino acids (isoform 1) and 479 amino acids (isoform 2).
The NADP+-dependent mitochondrial enzyme is called malic enzyme 3 and is encoded by the ME3 gene. The ME3 gene is located on chromosome 11q14.2 and is composed of 23 exons that generate five alternatively spliced mRNAs that collectively two protein isoforms, a 604 amino acid protein (isoform 1) and a 559 amino acid protein (isoform 2).
The role of the mitochondrial malic enzymes is principally to provide the cell with an alternate source of pyruvate under conditions where glycolytic flux in reduced. In these circumstances, the pyruvate generated by the actions of ME2 and/or ME3 comes from fumarate precursors such as glutamine.
In neurons, as well as in numerous types of tumor cells, mitochondrial malic enzymes allow for the utilization of the amino acid glutamine as a fuel source. When glutamine is de-aminated by glutaminase the resulting glutamate can also be de-aminated by glutamate dehydrogenase yielding 2-oxoglutarate (α-ketoglutarate) which can then be shunted to malate synthesis in the TCA cycle. The malate can then be decarboxylated to pyruvate via mitochondrial malic enzyme. The pyruvate can then be decarboxylated by the PDHc and the resulting acetyl-CoA can enter the TCA cycle ultimately allowing for glutamine carbons to be oxidized for ATP synthesis.
Within β-cells of the pancreas, this process driven by mitochondrial malic enzyme serves as an important means for the use of amino acid carbon oxidation for the stimulated secretion of insulin. Indeed, this process is energetically equal to glucose-stimulated insulin secretion (GSIS).
The malic enzyme encoded by the ME2 gene also plays a critical role in the progression of certain types of cancers, particularly leukemias. The role of ME2 in these types of cancers relates to its functions in mitochondrial biogenesis.
Reactions of Fatty Acid Synthesis
One might predict that the pathway for the synthesis of fatty acids would be the reversal of the oxidation pathway. However, this would not allow distinct regulation of the two pathways to occur even given the fact that the pathways are separated within different cellular compartments. The pathway for fatty acid synthesis occurs in the cytoplasm, whereas, oxidation occurs in the mitochondria. The other major difference is the use of nucleotide co-factors. Oxidation of fats involves the reduction of FAD and NAD+. Synthesis of fats involves the oxidation of NADPH. However, the essential reaction chemistry of the two processes are reversals of each other. Both oxidation and synthesis of fats utilize an activated two carbon intermediate, acetyl-CoA. However, the activated form of acetyl-CoA in fat synthesis exists temporarily bound to the enzyme complex as malonyl-CoA.
Acetyl-CoA Carboxylase (ACC) Reaction
The synthesis of malonyl-CoA is the first committed step of fatty acid synthesis and the enzyme that catalyzes this reaction, acetyl-CoA carboxylase (ACC), is the rate-limited enzyme and major site of regulation of fatty acid synthesis. Like other enzymes that transfer CO2 to substrates, ACC requires a biotin as a co-factor. Acetyl-CoA carboxylase is called an ABC enzyme due to the requirements for ATP, Biotin, and CO2 for the reaction.
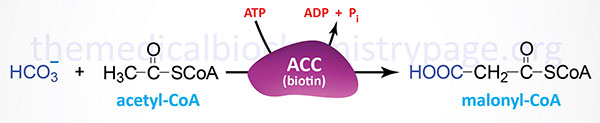
Humans express two major isoforms of ACC. These are identified as ACC1 (also called ACCα) and ACC2 (also called ACCβ).
The ACC1 enzyme is encoded by the ACACA gene. The ACACA gene is located on chromosome 17q12 and is composed of 66 exons that undergo alternative splicing to yield five splice variant mRNAs that generate four different precursor proteins from 2268 to 2383 amino acids in length. Transcriptional regulation of ACACA is effected by three promoters (PI, PII, and PIII), which are located upstream of exons 1, 2, and 5A, respectively. The PI promoter is a constitutive promoter, the PII promoter is regulated by various hormones, and the PIII promoter is expressed in a tissue-specific manner. The presence of the alternatively spliced exons does not alter the translation of the ACC1 protein which starts from an AUG present in exon 5.
The ACC2 enzyme is encoded by the ACACB gene. The ACACB gene is located on chromosome 12q24.11 and is composed of 58 exons that encode a precursor protein of 2,458 amino acids. The highest levels of expression of the ACACB gene are found in adipose tissue followed by heart and skeletal muscle.
Fatty Acid Synthase (FAS) Reactions
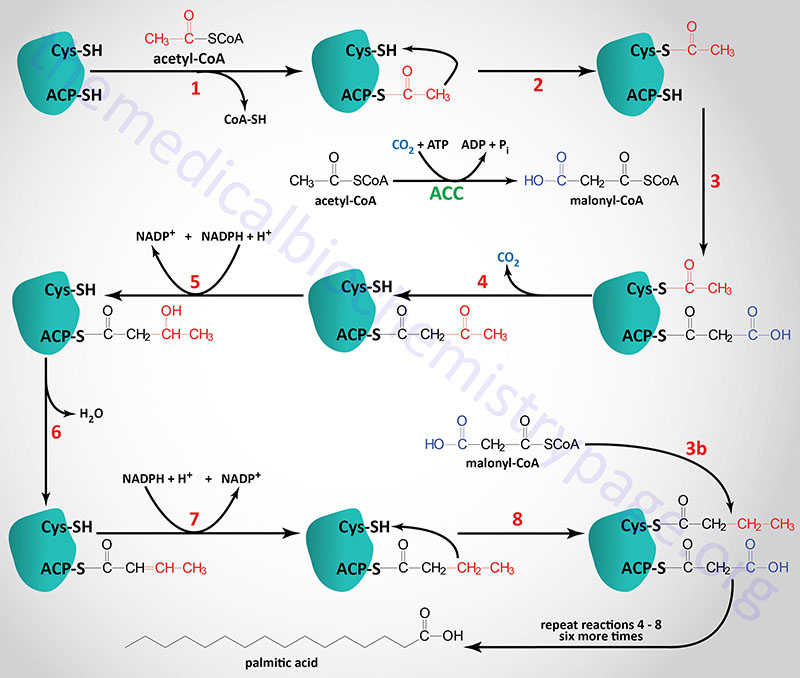
The acetyl groups that are the products of fatty acid and amino acid oxidation are linked to CoASH. As outlined in the Vitamins: Water and Fat Soluble page, CoASH contains a phosphopantetheine group coupled to AMP. The carrier of acetyl groups (and elongating acyl groups) during fatty acid synthesis is also a phosphopantetheine prosthetic group, however, it is attached to a serine hydroxyl in one of the active sites of the fatty acid synthase (FAS) complex. The carrier portion of the synthetic complex is called acyl carrier protein, ACP. This is somewhat of a misnomer in eukaryotic fatty acid synthesis since the ACP portion of the synthetic complex is simply one of many catalytic domains of a single polypeptide. The acetyl-CoA and malonyl-CoA are transferred to ACP by the action of the malonyl/acetyltransferase activity (also called acetyl-CoA transacylase and malonyl-CoA transacylase). The attachment of these carbon atoms to ACP allows them to enter the fatty acid synthesis cycle.
The synthesis of fatty acids from acetyl-CoA and malonyl-CoA is carried out by fatty acid synthase, FAS. Fatty acid synthase is encoded by the FASN gene. The FASN gene is located on chromosome 17q25.3 and is composed of 43 exons that encode a protein of 2511 amino acids. The highest level of expression of the FASN gene is found in adipose tissue at levels that are several hundred-fold higher than any other tissue.
The active FAS enzyme functions as a head-to-tail homodimer. All of the reactions of fatty acid synthesis are carried out by the multiple enzymatic activities of FAS. Like fat oxidation, fat synthesis involves four primary enzymatic activities. These are (in order of reaction), β-ketoacyl-ACP synthase (contained in a domain composed of amino acids 2–404), β-ketoacyl-ACP reductase (contained in a domain composed of amino acids 1876–2112), 3-hydroxyacyl-ACP dehydratase (contained in a domain composed of amino acids 874–1106) and enoyl-CoA reductase (contained in a domain composed of amino acids 1564–1854). The two reduction reactions require NADPH oxidation to NADP+. The domain that is required for attachment and transfer of acetyl-CoA and malonyl-CoA (acyltransferase domain) is composed of amino acids 493–810. The phosphopantetheine arm of FAS is attached to a domain composed of amino acids 2111-2179.
The primary fatty acid synthesized by FAS is the fully saturated 16 carbon fatty acid, palmitic acid (palmitate). Palmitate is then released from the enzyme via the thioesterase activity of FAS (contained in a domain composed of amino acids 2242–2488). Once released, palmitate can then undergo separate elongation and/or unsaturation to yield other fatty acid molecules.
Regulation of Fatty Acid Metabolism
Regulation by Acetyl-CoA Carboxylase: ACC
One must consider the body’s global integrated energy requirements in order to effectively understand how the synthesis and degradation of fats (and also carbohydrates) needs to be exquisitely regulated. The blood is the carrier of triglycerides [contained within very low density lipoproteins (VLDL) and chylomicrons], fatty acids bound to albumin, amino acids, lactate, ketone bodies, and glucose. The pancreas is the primary organ involved in sensing dietary and energy states primarily via glucose concentrations in the blood but also via glutamine levels and to a lesser extent fatty acid levels. In response to low blood glucose, glucagon is secreted, whereas, in response to elevated blood glucose insulin is secreted.
The regulation of fat metabolism occurs via two distinct mechanisms. One is short-term regulation which is regulation effected by events such as substrate availability, allosteric effectors and/or enzyme modification. The other is long-term regulation which involves the regulation of gene expression.
As indicated above, ACC represents the rate-limiting reaction of de novo fatty acid synthesis and humans express two forms of the enzyme identified as ACC1 and ACC2.
ACC1 is strictly cytosolic and is enriched in liver, adipose tissue, and lactating mammary tissue. ACC2 was originally discovered in rat heart but is also expressed in liver and skeletal muscle.
ACC2 has an N-terminal extension that contains a mitochondrial targeting motif and is found associated with carnitine palmitoyltransferase 1 (CPT1) allowing for rapid regulation of CPT1 by the malonyl-CoA produced by ACC2. The localization of ACC2 and the inhibition of CPT1 by its product, malonyl-CoA, indicates that ACC2 is primarily involved in the regulation of fatty acid oxidation as opposed to fatty acid synthesis.
Both isoforms of ACC are allosterically activated by citrate and inhibited by palmitoyl-CoA and other short- and long-chain fatty acyl-CoAs. Citrate triggers the polymerization of ACC1 which leads to significant increases in its activity. Although ACC2 does polymerize in response to citrate, the extent of oligomerization does not appear to be as great as for ACC1, which is presumed to be due to its mitochondrial association. Glutamate and other dicarboxylic acids can also allosterically activate both ACC isoforms.
ACC activity can also be affected by phosphorylation. Both ACC1 and ACC2 contain at least eight sites that undergo phosphorylation. The sites of phosphorylation in ACC2 have not been as extensively studied as those in ACC1. Phosphorylation of ACC1 at three serine residues (S79, S1200, and S1215) by AMPK leads to inhibition of the enzyme. AMPK also phosphorylates and inhibits the activity of ACC2. Glucagon-stimulated increases in cAMP and subsequently to increased PKA activity also lead to phosphorylation of ACC where ACC2 is a better substrate for PKA than is ACC1. Epinephrine activation of β-adrenergic receptors also results in increased PKA activity and consequently phosphorylation and inhibition of ACC1.
The activating effects of insulin on ACC are complex and not completely resolved. It is known that insulin leads to the dephosphorylation of the phosphoserines in ACC1 that are AMPK targets in the heart enzyme. This insulin-mediated effect has not been observed in hepatocytes or adipose tissues cells. At least a portion of the activating effects of insulin are related to changes in cAMP levels. Early evidence had shown that phosphorylation and activation of ACC occurs via the action of an insulin-activated kinase. However, contradicting evidence indicates that although there is insulin-mediated phosphorylation of ACC this does not result in activation of the enzyme.
Regulation of ACC2 by Prolyl Hydroxylation
The activity of ACC2 is also regulated by hydroxylation. As the level of glucose rises the extent of ACC2 hydroxylation increases and the level of malonyl-CoA rises resulting in inhibition of fatty acid oxidation. The primary enzyme responsible for the hydroxylation of ACC2 is a member of the prolyl hydroxylase domain (PHD) family of enzymes. Humans express three PHD family member encoding genes, PHD1, PHD2, and PHD3. The PHD family enzymes are members of the large family of 2-oxoglutarate (α-ketoglutarate) and Fe2+ (iron)-dependent dioxygenases (2OG-oxygenases).
Within cardiac and skeletal muscle the PHD3 encoded enzyme has been shown to hydroxylate ACC2. When AMPK phosphorylates ACC2 during periods of energetic stress, PHD3 can no longer hydroxylate the enzyme. Studies have shown that loss of PHD3 activity (in knock-out mice) results in a significant reduction in ACC2 activity. The reduced ACC2 activity, resulting in reduced levels of malonyl-CoA, is associated with increased fatty acid delivery to the mitochondria for oxidation which promotes exercise capacity.
Regulation by Malonyl-CoA Decarboxylase: MCD
The intracellular levels of malonyl-CoA represent a balance between its synthesis from acetyl-CoA by ACC and its utilization in fatty acid synthesis by FAS as well as by its degradation to acetyl-CoA via the action of malonyl-CoA decarboxylase (MCD). Indeed, MCD is involved in regulating malonyl-CoA levels in multiple tissues. MCD is found in the cytosol, the mitochondria, and in peroxisomes.
The MCD enzyme is encoded by the MLYCD gene. The MLYCD gene is located on chromosome 16q23.3 and is composed of 5 exons encoding a protein of 493 amino acids.
Inhibition of MCD results in reduced rates of fatty acid oxidation in highly oxidative tissues such as the heart. As well, MCD inhibition leads to reduced triglyceride content in lipid synthesizing tissues such as the liver.
When hypothalamic MCD levels are experimentally increased in laboratory animals there is a dramatic increase in food intake, weight gain, and ultimately results in obesity. Conversely, inactivation of the MCD gene in mice protects the animals from high-fat diet-induced insulin resistance. Additionally, chemical inhibition of MCD leads to reduced macrophage-associated inflammation in conditions of insulin resistance. The primary mechanism for this effect appears to be via the accumulation of long-chain fatty acids which in turn activate PPARα.
Interestingly, reduced MCD activity also exerts a beneficial effect in the hypothalamus. Transcriptional regulation of the MCD gene is exerted by PPARα, a major transcription factor involved in the regulation of fatty acid oxidation. Hypothalamic PPARα has been shown to play a role in the regulation of appetite, presumably via enhanced expression of MCD. PPARα-mediated increases in MCD levels results in reduced levels of malonyl-CoA in the hypothalamus. This is associated with increased food intake in the FAS knock-out mice demonstrating that malonyl-CoA levels are indeed responsible for the hypophagic effects observed in the FAS knock-out mice. The potential therapeutic benefits to reduced MCD activity in the treatment of obesity and diabetes are currently undergoing intensive investigation.
Additional Processes Regulating Fatty Acid Synthesis
Control of a given pathways’ regulatory enzymes can also occur by alteration of enzyme synthesis and turn-over rates. These changes are long term regulatory effects. Insulin stimulates ACC and FAS synthesis, whereas, starvation leads to decreased synthesis of these enzymes. Adipose tissue lipoprotein lipase levels also are increased by insulin and decreased by starvation. However, in contrast to the effects of insulin and starvation on adipose tissue, their effects on heart lipoprotein lipase are just the inverse. This allows the heart to absorb any available fatty acids in the blood in order to oxidize them for energy production. Starvation also leads to increases in the levels of fatty acid oxidation enzymes in the heart as well as a decrease in FAS and related enzymes of synthesis.
Adipose tissue contains hormone-sensitive lipase (HSL), that is activated by PKA-dependent phosphorylation leading to increased fatty acid release to the blood. The activity of HSL is also affected through the action of AMPK, however, AMPK-mediated phosphorylation of HSL is inhibitory. The phosphorylation and inhibition of HSL by AMPK may seem paradoxical since the release of fatty acids stored in triglycerides would seem necessary to promote the production of ATP via fatty acid oxidation and the major function of AMPK is to shift cells to ATP production from ATP consumption. This paradigm can be explained if one considers that if the fatty acids that are released from triglycerides are not consumed they will be recycled back into triglycerides at the expense of ATP consumption. Thus, it has been proposed that inhibition of HSL by AMPK mediated phosphorylation is a mechanism to ensure that the rate of fatty acid release does not exceed the rate at which they are utilized either by export or oxidation.
In the liver the net result of activation of HSL (due to increased acetyl-CoA levels) is the production of ketone bodies. This would occur under conditions where insufficient carbohydrate stores and gluconeogenic precursors were available in liver for increased glucose production. The increased fatty acid availability in response to glucagon or epinephrine is assured of being completely oxidized since both PKA and AMPK also phosphorylate (and as a result inhibits) ACC, thus inhibiting fatty acid synthesis.
Insulin, on the other hand, has the opposite effect to glucagon and epinephrine leading to increased glycogen and triglyceride synthesis. One of the many effects of insulin is to lower cAMP levels which leads to increased dephosphorylation through the enhanced activity of protein phosphatases such as PP-1. With respect to fatty acid metabolism this yields dephosphorylated and inactive hormone sensitive lipase. Insulin also stimulates certain phosphorylation events. This occurs through activation of several cAMP-independent kinases.
Regulation of fat metabolism also occurs through malonyl-CoA induced inhibition of carnitine acyltransferase 1 (CPT1). This functions to prevent the newly synthesized fatty acids from entering the mitochondria and being oxidized. Indeed, as pointed out above, the localization of ACC2 to the mitochondrial outer membrane allows for rapid inhibition of CPT1 when fatty acid synthesis has been stimulated.
Mitochondrial Fatty Acid Synthesis
Synthesis of fatty acids in the mitochondria plays a critical role in the assembly of the components of the oxidative phosphorylation machinery as well as playing a role in the processing of mitochondrial RNAs. The process of mitochondrial fatty acid synthesis closely resembles a bacterial type II fatty acid synthesis system (FASII) that involves individual enzymes for each step in the pathway. This is unlike cytosolic fatty acid synthesis which is catalyzed by two enzymes, acetyl-CoA carboxylase 1 (ACC1) and fatty acid synthase (FAS).
Initiation of mitochondrial fatty acid synthesis involves the use of malonic acid. Malonic acid is most likely transported into the mitochondria via the carnitine transport process similar to that required for mitochondrial entry of long-chain fatty acids in the process of fatty acid β-oxidation.
Mitochondrial fatty acid synthesis requires an acyl-carrier protein to which a 4′-phosphopantetheine (4′-PP) co-factor
has been added to a conserved Ser residue. This mitochondrial ACP function is similar to the ACP portion of cytosolic fatty acid synthase (FAS). The mitochondrial ACP is a function of a subunit of mitochondrial oxidative phosphorylation complex I and is encoded by the NDUFAB1 (NADH:ubiquinone oxidoreductase subunit AB1) gene. An enzyme that is poorly characterized in humans, phosphopantetheinyl transferase, attaches the 4′-PP co-factor to the appropriate Ser residue in the NDUFAB1 encoded protein. This modified NDUFAB1 protein is the scaffold upon which mitochondrial fatty acids are synthesized.
The first enzyme involved in mitochondrial fatty acid synthesis is acyl-CoA synthetase family member 3 that is encoded by the ACSF3 gene. The ACSF3 encoded enzyme is also referred to as malonyl-CoA synthetase given that it exhibits specificity for malonic acid and methylmalonic acid. The major product of the ACSF3 encoded enzyme is malonyl-CoA.
The ACSF3 gene is located on chromosome 16q24.3 and is composed of 16 exons that generate four alternatively spliced mRNAs that collectively encode two different precursor proteins of 576 amino acids (isoform 1) and 311 amino acids (isoform 2). Translation of the isoform 2 protein initiates at a downstream start codon relative to isoform 1.
Mutations in the ACSF3 gene result in the disorder called combined malonic and methylmalonic aciduria (CMAMMA). Symptoms of CMAMMA commonly begin in early childhood and include ketoacidosis, hypoglycemia, muscle issues (dystonia and hypotonia), developmental delay, failure to thrive, and potentially coma. In some individuals symptoms don’t appear until adulthood and include seizures, loss of memory, difficulty thinking, or psychiatric abnormalities.
The malonyl-CoA produced in the first reaction of mitochondrial fatty acid synthesis is the substrate for malonyl-CoA-acyl carrier protein transacylase which is encoded by the MCAT gene. The MCAT enzyme is exclusively localized to the mitochondria. MCAT attaches the malonyl-CoA to the mitochondrial ACP, generating malonyl-ACP.
The MCAT gene is located on chromosome 22q13.2 and is composed of 4 exons that generate two alternatively spliced mRNAs encoding two distinct precursor proteins. The isoform a precursor is 390 amino acids and the isoform b precursor is 180 amino acids.
Malonyl-ACP is successively elongated in the next series of reactions that begins through the action of 3-oxoacyl-ACP synthase that is encoded by the OXSM gene. This enzyme catalyzes a condensation between the malonyl-ACP and an acyl-ACP, which would be acetyl-ACP in the first reaction of elongation. The condensation reaction releases CO2 and generates an acyl-ACP of an even number of carbon atoms. The mechanism by which the initial acetyl-ACP is generated is unclear. The OXSM encoded enzymes carries out each subsequent round of elongation where the substrates are the acyl-ACP product and a malonyl-ACP.
The OXSM gene is located on chromosome 3p24.2 and is composed of 3 exons that generate two alternatively spliced mRNAs encoding two distinct precursor proteins of 459 amino acids (isoform 1) and 376 amino acid (isoform 2).
The 3-ketoacyl-ACP product of the OXSM encoded enzyme is the substrate for mitochondrial 3-ketoacyl-ACP reductase. Mitochondrial 3-ketoacyl-ACP reductase is a heterotetrameric complex (α2β2) where the α-subunits are 17β-hydroxysteroid dehydrogenase type 8 and the β-subunits are carbonyl reductase type 4. The product of the 3-ketoacyl-ACP reductase is a 3R-hydroxyacyl-ACP. The 17β-hydroxysteroid dehydrogenase type 8 is encoded by the HSD17B8 gene and the carbonyl reductase type 4 is encoded by the CBR4 gene. The HSD17B8 encoded enzyme is also involved in the inactivation of estradiol, testosterone, and dihydrotestosterone.
The HSD17B8 gene is located on chromosome 6p21.32 and is composed of 9 exons that encode a 261 amino acid protein.
The CBR4 gene is located on chromosome 4q32.3 and is composed of 9 exons that encode a 237 amino acid protein. The CBR4 encoded enzyme is a member of the large family of short-chain dehydrogenases/reductases (SDR).
The 3R-hydroxyacyl-ACP is then converted to a trans-2-enoyl-ACP via the action of hydroxyacyl-thioester dehydratase type 2 that is encoded by the HTD2 gene. The HTD2 gene is located on chromosome 3p14.3 and is composed of 7 exons that generate four alternatively spliced mRNAs that differ in their 5′-UTR but that encode the same 168 amino acid protein.
The HTD2 gene is located downstream of the gene for ribonuclease P/MRP subunit p14 (RPP14) and these two genes appear to be co-transcribed into a bicistronic mRNA that may contain both open reading frames.
The last step in mitochondrial fatty acid synthesis reduces the trans-2-enoyl-ACP to an acyl-ACP in an NADPH-dependent reaction catalyzed by mitochondrial trans-2-enoyl-CoA reductase encoded by the MECR gene. Mitochondrial trans-2-enoyl-CoA reductase is a member of the medium-chain alcohol dehydrogenase/reductase (MDR) family.
The MECR gene is located on chromosome 1p35.3 and is composed of 18 exons that generate nine alternatively spliced mRNAs that collectively encode five distinct protein isoforms.
Mutations in the MECR gene are associated with hypotonia, dystonia, spasticity, optic atrophy, and abnormalities in brain development, particularly in the basal ganglia.
The acyl-ACP product of the mitochondrial trans-2-enoyl-CoA reductase can then be elongated by serving as the substrate for the OXSM encoded enzyme. When the eight carbon saturated fatty acid octanyl-ACP product is generated there are two fates, one is diversion of the octanoyl moiety into the lipoic acid synthesis pathway, the other is further elongation to either myristoyl-ACP (C14) or palmitoyl-ACP (C16).
The mitochondrial acyl carrier protein, although being a subunit of complex I of the oxidative phosphorylation pathway, is also critical for the assembly of other complexes of oxidative phosphorylation. Mitochondrial ACP interacts with proteins of the LYRM (Leucine-tYrosine-aRginine Motif) family. Individual LYRM proteins not only serve as accessory subunits or assembly factors of complexes I, II, III, and IV, they serve as assemble or accessory subunits of mitochondrial ribosomes, they are essential for the biogenesis of Fe-S cluster proteins/complexes, and they are also involved in the metabolism of acetate.
Humans express ten genes that encode LYRM family proteins. These genes are LYRM1, LYRM2, LYRM4, LYRM7, LYRM9, NDUFB8 (LYRM3), ETFRF1 (LYRM5), NDUFA6 (LYRM6), SDHAF1 (LYRM8), and SDHAF3 (LYRM10). NDUFB8 encodes NADH:ubiquinone oxidoreductase subunit B9. ETFRF1 encodes electron transfer flavoprotein regulatory factor 1. NDUFA6 encodes NADH:ubiquinone oxidoreductase subunit A6. SDHAF1 and SDHAF3 encode succinate dehydrogenase complex assembly factor 1 and 3, respectively.
Mitochondrial Synthesis of Lipoic Acid
Alpha-lipoic acid, LA, (chemical name: 1,2-dithiolane-3-pentanoic acid; also known as thioctic acid), is a naturally occurring dithiol compound synthesized enzymatically in the mitochondria. The initial substrate for lipoic acid synthesis is the octanoic acid attached to the ACP sulfhydryl of the mitochondrial fatty acid synthesis enzyme, malonyl-CoA-acyl carrier protein transacylase which, as indicated above, is encoded by the MCAT gene. The octanoyl group is transferred to the H-protein of the glycine cleavage complex via the action of lipoyl (octanoyl) transferase 2 (encoded by the LIPT2 gene). Lipoic acid synthase (encoded by the LIAS gene) then inserts sulfur atoms into the octanoyl group on the H-protein. Finally, lipoyltransferase 1 (encoded by the LIPT1 gene) transfers the lipoyl group from the H-protein to E2 subunits of several enzyme complexes that includes the pyruvate dehydrogenase complex (PDHc), the 2-oxoglutarate (α-ketoglutarate) dehydrogenase complex (OGDH), and the branched-chain keto acid dehydrogenase complex (BCKD).
The LIPT2 gene is located on chromosome 11q13.4 and is composed of 2 exons that as a result of the use of alternative splice sites generates three mRNAs, each of which encode distinct protein isoforms.
The LIAS gene is located on chromosome 4p14 and is composed of 11 exons that generate six alternatively spliced mRNAs, each of which encodes a distinct protein isoform.
The LIPT1 gene is located on chromosome 2q11.2 and is composed of 7 exons that generate five alternatively spliced mRNAs, all of which encode the same 373 amino acid precursor protein.
Mutations in the LIPT2, LIAS, and LIPT1 genes have been identified in humans and they result in defects in lipoic acid synthesis. Defective lipoic acid synthesis is associated with neonatal-onset epilepsy, defective mitochondrial
energy metabolism, and hyperglycinemia.
ChREBP: Master Lipid Regulator in the Liver
When glycogen stores are maximal in the liver, excess glucose is diverted into the lipid synthesis pathway as a direct result of the mechanisms activated by insulin in this organ. Glucose is catabolized to acetyl-CoA and the acetyl-CoA is used for de novo fatty acid synthesis. The fatty acids are then incorporated into triglycerides and exported from hepatocytes as very-low-density lipoproteins (see the Lipoproteins, Blood Lipids, and Lipoprotein Metabolism page for more details) and ultimately stored as triglycerides in adipose tissue.
A diet rich in carbohydrates leads to stimulation of both the glycolytic and lipogenic pathways. The transcription rates of genes encoding glucokinase (GCK) and liver pyruvate kinase (PKLR) of glycolysis and ATP-citrate lyase (ACLY), ACC1 (ACACA), and FAS (FASN) of lipogenesis are modulated in response to increased carbohydrate intake. In addition, the enzymes encoded by these genes are subject to post-translational and allosteric regulation. These genes contain glucose- or carbohydrate-response elements (ChoRE) that are responsible for their transcriptional regulation.
One transcription factor that exerts control over glucose and lipid homeostasis is sterol-response element-binding protein (SREBP), in particular SREBP-1c. The regulation and actions of SREBP are discussed in the Cholesterol: Synthesis, Metabolism, and Regulation page. SREBP controls the expression of a number of genes involved in lipogenesis and its own transcription is increased by insulin and repressed by glucagon. However, SREBP activity alone cannot account for the stimulation of glycolytic and lipogenic gene expression in response to a carbohydrate rich diet.
A search for potential additional regulatory factors revealed a basic helix-loop-helix/leucine zipper (bHLH/LZ) transcription factor which was identified as carbohydrate-responsive element-binding protein (ChREBP). ChREBP is most highly expressed in the liver but is also abundantly expressed in white adipose tissue (WAT), brown adipose tissue (BAT), pancreatic β-cells, small intestine, kidney, adrenal glands, and brain. ChREBP was identified as a major glucose-responsive transcription factor and it is required for glucose-induced expression of PKLR gene, which encodes L-PK, and the lipogenic genes encoding ACC1 and FAS.
ChREBP is encoded by the MLXIPL (MLX interacting protein like) gene. The MLXIPL gene is located on chromosome 7q11.23 and is composed of 19 exons that generate four alternatively spliced mRNAs that encode isoforms alpha (α), beta (β), gamma (γ), and delta (δ). Deletion of several genes, including MLXIPL, is the cause of Williams-Beuren syndrome. In skeletal muscle, the ChREBP homolog is called MondoA and it is encoded by the MLXIP gene.
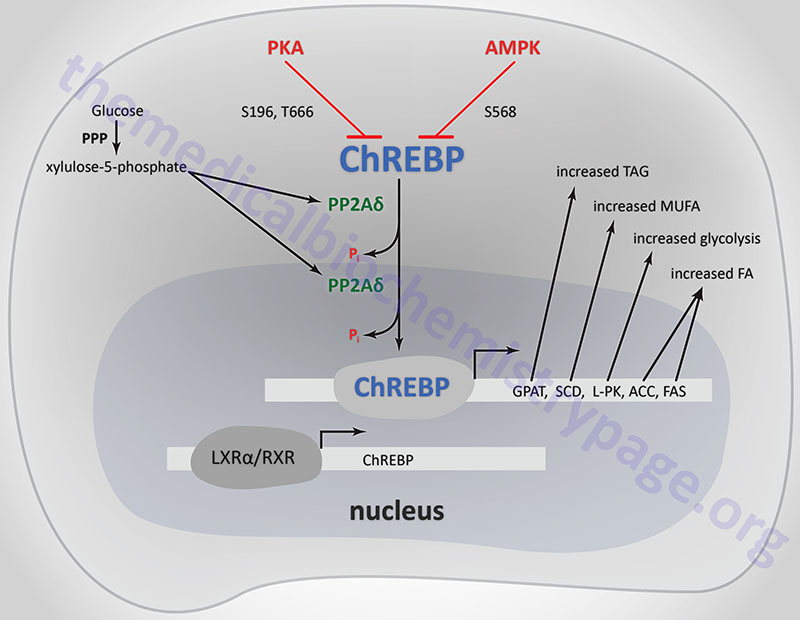
ChREBP does not bind to ChoREs as a typical homodimeric bHLH transcription factor. ChREBP interacts with another bHLH protein identified as MAX-like protein X (MLX). MLX is a member of the MYC/MAX/MAD family of transcription factors that serve as interacting partners in transcription factor networks. The binding of MLX to ChREBP occurs within a domain located in the C-terminal portion of ChREBP. This interaction between ChREBP and MLX is essential for DNA-binding.
Regulation of ChREBP (MLXIPL) Expression
The liver X receptors (LXR) are members of the steroid/thyroid hormone superfamily of nuclear receptors that regulate gene expression by binding to specific target DNA sequences. There are two forms of the LXRs, LXRα and LXRβ. The LXR form heterodimers with the retinoid X receptors (RXR) and as such can regulate gene expression either upon binding oxysterols (e.g. 22R-hydroxycholesterol) or 9-cis-retinoic acid. LXR are also important regulators of the lipogenic pathway.
Recent evidence has demonstrated that the MLXIPL gene is a direct target of LXR and that glucose itself can bind and activate LXRs. There are two LXR binding sites in the promoter region of the MLXIPL gene. Within the liver the thyroid hormone receptor beta (TRβ), a member of the nuclear receptor family, also binds to and activates expression of the MLXIPL gene.
Regulation of ChREBP Activity
When glucose levels rise, protein phosphatase 2A delta (PP2Aδ) is activated by xylulose 5-phosphate which is a product of glucose metabolism in the pentose phosphate pathway. PP2Aδ dephosphorylates S196 resulting in translocation of ChREBP into the nucleus. In the nucleus PP2Aδ dephosphorylates T666 which allows ChREBP to bind to specific sequence elements (ChoRE) in target genes. Other glucose-derived metabolites, including glucose 6-phosphate and acetyl-CoA, are potential activators of ChREBP translocation into the nucleus.
The activity of ChREBP is regulated by post-translational modifications as well as sub-cellular localization. The kinases PKA and AMPK both phosphorylate ChREBP rendering it inactive as a transcriptional activator. PKA is known to phosphorylate ChREBP on serine 196 (S196) and threonine 666 (T666), whereas, AMPK phosphorylates ChREBP at serine 568 (S568). Under conditions of low (basal) glucose concentration, ChREBP is phosphorylated and resides in the cytosol.
Subsequent to the identification of glucose as an enhancer of ChREBP activity it was found that fructose is a more potent activator than glucose. Fructose-mediated activation of ChREBP involves acetylation of the protein and increased DNA binding to ChREBP target genes.
Additional mechanisms of glucose-mediated regulation of ChREBP activity were made apparent when it was shown that mutations in the PKA phosphorylation sites (S196 and T666) did not completely abolish glucose-responsiveness. Further analysis of ChREBP regulation in response to glucose administration was shown to be due to domains present in the amino terminal portion of ChREBP. The glucose sensing domain (GSM) is actually composed of two distinct sub-domains identified as the low-glucose inhibitory domain (LID) and the glucose-responsive activation conserved element (GRACE). Under conditions of low glucose the LID inhibits transcriptional transactivation by the GRACE domain. This inhibition is reversed by glucose or a glucose metabolite such as glucose-6-phosphate or acetyl-CoA.
Recently a newly discovered mechanism of regulated ChREBP activity involves the tissue-specific transcription of an alternatively spliced form of ChREBP. This alternative splice variant contains a novel upstream exon (identified as exon 1b) and bypasses the originally identified exon 1 (now identified as exon 1a). This novel mechanism of ChREBP activity has been shown to occur in adipose tissue and represents a potent mechanism for glucose-mediated modulation of adipose tissue fatty acid synthesis and insulin sensitivity.
The original ChREBP is now referred to as ChREBP-α and the novel alternative splice form is called ChREBP-β. The means by which glucose plays a role in adipose tissue ChREBP activity is that glucose, or a metabolite, activates the transcriptional activity of ChREBP-α by the mechanisms described earlier. One of the important adipose tissue targets for glucose-activated ChREBP-α is the upstream transcriptional activation site that regulates the transcription of ChREBP-β. Following adipose tissue activation of ChREBP-β expression, both ChREBP-α and ChREBP-β work in concert to dramatically alter lipogenic gene expression.
Regulation of ChREBP Activity by O-GlcNAcylation
In addition to phosphorylation by PKA and AMPK and fructose-mediated acetylation, ChREBP activity is regulated by O-GlcNAcylation. The O-GlcNAcylation of ChREBP results in increased stabilization of the ChREBP protein and increased transcriptional activity.
Additional mechanisms of ChREBP activity are the result of its O-GlcNAcylation. Upon O-GlcNAcylation, ChREBP interacts with host cell factor 1 (encoded by the HCFC1 gene) which increases the recruitment of O-GlcNAc transferase (OGT) to ChREBP. In addition, the HCF1–ChREBP complex is involved in the recruitment of the histone demethylase, PHF2 (PHD finger protein 2 where PHD refers to plant homeodomain), resulting in the epigenetic modification of the promoter region of several lipogenic genes. PHF2 is a member of the Jumonji C domain-containing (JmjC) histone demethylase family of enzymes.
ChREBP Target Genes
Genes whose patterns of expression are under the control of ChREBP activity are involved in glycolysis, the pentose phosphate pathway, gluconeogenesis, and lipogenesis.
Genes that are involved in glucose metabolism that are regulated by ChREBP include those encoding liver pyruvate kinase (L-PK encoded by the PKLR gene), glucokinase regulatory protein (GCKR), and GLUT2 (SLC2A2).
Genes encoding enzymes in the gluconeogenesis pathway that are subject to regulation by ChREBP such as G6PC1, which encodes the predominant form of glucose-6-phosphatase, and FBP1 which encodes the liver form of fructose-1,6-bisphosphatase.
Genes encoding enzymes involved in the pentose phosphate pathway that are regulated by ChREBP include those encoding glucose-6-phosphate dehydrogenase (G6PD) and transketolase (TKT).
ChREBP also regulates the expression of several genes involved in the metabolism of fructose in the small intestines and the liver. These genes include SLC2A2 and SLC2A5 which encode GLUT2 and GLUT5, respectively, KHK which encodes ketohexokinase (commonly referred to as fructokinase), ALDOB which encodes aldolase B, and TKFC which encodes triokinase and FMN cyclase.
Within adipose tissue ChREBP-α and ChREBP-β function together to dramatically increase the transcription of lipogenic genes in this tissue such as FASN (fatty acid synthase) and ACACA (acetyl-CoA carboxylase 1, ACC1). In addition, it has been shown that when expression of MLXIPL is reduced the expression levels of the genes encoding glycerol 3-phosphate acyltransferases (GPAM, GPAT2, GPAT3, GPAT4) and Δ9-stearoyl-CoA desaturase (SCD) are also reduced.
The glycerol 3-phosphate acyltransferases esterify glycerol-3-phospate generating lysophosphatidic acid (LPA) which is the first step in the synthesis of triglycerides (TG; or triacylglycerides, TAG) as described below.
SCD is the rate-limiting enzyme involved in the synthesis of the major monounsaturated fatty acids oleic acid (18:1) and palmitoleic acid (16:1) as described below in the section on Elongation and Desaturation.
Elongation and Desaturation
Elongation
The fatty acid product released from FAS is palmitate (via the action of palmitoyl thioesterase) which is a 16:0 fatty acid (i.e. 16 carbons and no sites of unsaturation). Elongation of fatty acids occurs in the cytosol, the mitochondria, and the endoplasmic reticulum (microsomal membranes). The predominant site of fatty acid elongation is in the ER membranes.
Elongation involves condensation of fatty acyl-CoA groups with either acetyl-CoA or malonyl-CoA. The resultant product is two carbons longer (CO2 is released from malonyl-CoA as in the FAS reaction) which undergoes reduction, dehydration and reduction yielding a saturated fatty acid extended in length by two carbon atoms. The reduction reactions of elongation require NADPH as co-factor.
Cytoplasmic fatty acid elongation is essentially an extension of the normal fatty acid synthesis reaction. Acetyl-CoA carboxylase (ACC) produces malonyl-CoA and then FAS utilizes acetyl-CoA and the malonyl-CoA to elongate the primary end-product of normal fatty acid synthesis, palmitate. Mitochondrial fatty acid elongation involves a fatty acyl-CoA and acetyl-CoA units and the enzyme trans-2-enoyl-CoA reductase (encoded by the MERC gene). The mitochondrial fatty acid elongation pathway is a minor pathway in humans.
The microsomal fatty acid elongation pathway represents the major pathway for the elongation of both endogenous and dietary fatty acids. The overall pathway involves four enzymes with the substrates being fatty acyl-CoAs, malonyl-CoA, and NADPH.
In the initial step in the pathway a 3-keto acyl-CoA synthase catalyzes the condensation of malonyl-CoA with a fatty acyl-CoA precursor. Humans express seven distinct 3-keto acyl-CoA synthase encoding genes. These seven genes are designated ELOVL1–ELOVL7 which stands for ELOngation of Very Long fatty acids. Fatty acyl-CoA substrate specificity and the rate of fatty acid elongation in the microsomes is determined by the ELOVL enzymes and not the reductases nor the dehydratase.
- The ELOVL1 encoded enzyme exhibits preference for saturated fatty acids in the 18-carbon (C18) to C26 length with highest activity towards the C22:0 saturated fatty acid called behenic acid.
- The ELOVL2 encoded enzyme elongates polyunsaturated fatty acids (PUFA) of C20 and C22 length with highest activity towards the C20:4 PUFA, arachidonic acid.
- The ELOVL3 encoded enzyme elongates saturated fatty acids with highest preference for stearic acid (C18:0).
- The ELOVL4 encoded enzyme preferentially elongates very long-chain saturated fatty acids such as lignoceric (C24:0) and cerotic (C26:0) acids. Critical enzyme in the synthesis of very long-chain PUFA (C26 to C38) and very long-chain saturated fatty acids. Mutations in the ELOVL4 gene are associated with inherited forms of macular degeneration.
- The ELOVL5 encoded enzyme elongates PUFA with highest preference for the C18:3 fatty acids, α-linolenic acid (ALA) and gamma(γ)-linolenoyl-CoA (GLA). ALA has sites of unsaturation at carbons 9, 12, and 15 and is the precursor fatty acid in the pathway of omega-3 PUFA synthesis. GLA has sites of unsaturation at carbons 6, 9, and 12 and is an intermediate in the pathway of arachidonic acid synthesis. ELOVL5 can also elongate some monounsaturated fatty acids, such as palmitoleic acid (C16:1).
- The ELOVL6 encoded enzyme elongates saturated fatty acids with 12, 14, and 16 carbon atoms with highest activity towards palmitic acid (C16:0).
- The ELOVL7 encoded enzyme has been shown to elongate a range of both saturated and unsaturated fatty acids such as the C16:0, C18:0, C18:1, C18:2, C18:3, and C20:4 fatty acids. The C18:3 substrates of ELOVL7 are the same as those used by the ELOVL5 enzyme. ELOVL7 shows no elongation activity towards fatty acids longer than 20 carbons.
The ELOVL genes are expressed in tissue specific patterns as well as being subjected to dietary and hormonal regulation. ELOVL1, ELOVL2, ELOVL3, ELOVL5, and ELOVL6 are highly expressed in the liver. The heart does not express ELOVL2 but does express ELOVL1, ELOVL5, and ELOVL6.
In the second reaction of fatty acid elongation a 3-ketoacyl-CoA reductase activity (also called 3-oxoacyl-CoA reductase) reduces the resulting 3-ketoacyl-CoA intermediate to a 3-hydroxyacyl-CoA. The 3-ketoacyl-CoA reductase activity of mammalian fatty acid elongation is associated with the enzyme hydroxysteroid 17-β dehydrogenase 12 encoded by the HSD17B12 gene.
The HSD17B12 gene is located on chromosome 11p11.2 and is composed of 18 exons that encode a 312 amino acid protein. Elongation of fatty acids in the mitochondria utilizes a related enzyme called 3-oxoacyl-ACP synthase that is encoded by the OXSM gene.
The third step in fatty acid elongation involves the dehydration of the 3-hydroxy species and it is catalyzed by 3-hydroxyacyl-CoA dehydratase 2 (encoded by the HACD2 gene). Humans express four genes of the HACD family (HACD1, DACD2, DACD3, and HACD4) but only the protein encode by HACD2 is involved in fatty acid elongation.
The HACD2 gene is located on chromosome 3q21.1 and is composed of 11 exons that generate five alternatively spliced mRNAs that collectively encode four protein isoforms.
In the fourth step of fatty acid elongation the product of HACD2 catalyzed reaction is reduced by trans-2,3-enoyl-CoA reductase, an member of the steroid 5-alpha reductase family. The trans-2,3-enoyl-CoA reductase is encoded by the TECR gene.
The TECR gene is located on chromosome 19p13.12 and is composed of 15 exons that generate two alternatively spliced mRNAs, both of which encode distinct protein isoforms.
The fatty acid elongases function in concert with the fatty acid desaturases (next section) to generate many of the long chain monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA) that are found in cellular lipids. For example ELOVL6 and stearoyl-CoA desaturase (encoded by the SCD gene) generate the 18-carbon monounsaturated fatty acid (C18:1), oleic acid. ELOVL2 and ELOVL5 function along with the delta-5 (D5D) and delta-6 (D6D) desaturases to generate the omega-6 PUFA, arachidonic acid (C20:4) and the omega-3 PUFA, docosahexaenoic acid, DHA (C22:6).
Desaturation
Like the elongation of fatty acids, the desaturation of fatty acids occurs in the ER membranes. In mammalian cells fatty acid desaturation involves three broad specificity fatty acyl-CoA desaturases (non-heme iron containing enzymes). These enzymes introduce unsaturation at C5, C6, or C9 of various fatty acid substrates. The names of these enzymes are Δ5-eicosatrienoyl-CoA desaturase (D5D), Δ6-oleoyl(linolenoyl)-CoA desaturase (D6D), and Δ9-stearoyl-CoA desaturase (SCD).
The D5D enzyme is encoded by the FADS1 gene located on chromosome 11q12.2 and is composed of 13 exons that encode a 501 amino acid protein. The FADS1 encoded enzyme has near exclusive activity towards PUFA with 20 carbon atoms.
The D6D enzyme is encoded by the FADS2 gene located on the same chromosome as the FADS1 gene, 11q12.2. The FADS2 gene is composed of 13 exons that generates three alternatively spliced mRNAs that encode three distinct isoforms of the enzyme. This chromosomal region is referred to as the FADS cluster and harbors an additional desaturase gene identified as FADS3. The FADS2 encoded enzyme desaturates linoleic acid (omega-6 18:2; also written 18:2n-6) generating γ-linolenic acid (GLA; 18:3n-6). FADS2 also desaturates the α-linolenic acid (omega-3 18:3; also written 18:3n-3) generating stearidonic acid (18:4n-3). FADS2 also functions as a Δ4 and a Δ8 desaturase. The Δ4 desaturase activity is functional on 22:4n-6 (adrenic acid) generating docosapentaenoic acid (22:5n-6; DPAn-6; trivial name: osbond acid) and on the omega-3 form of docosapentaenoic acid (22:5n-3; DPAn-3; trivial name clupanodonic acid) generating docosahexaenoic acid (22:6n-3). The Δ8 desaturase activity acts on 20:2n-6 (eicosadienoic acid) and 20:3n-3 (eicosatrienoic acid) generating 20:3n-6 (dihomo-γ-linolenic acid, DGLA) and 20:4n-3 (eicosatetraenoic acid), respectively.
The SCD enzyme is encoded by the SCD gene (also known as FADS5) that is located on chromosome 10q24.31 and is composed of 6 exons that generate at least two mRNAs that differ by alternative polyadenylation signal sequence utilization. Both SCD mRNAs encode the same 359 amino acid protein.
The electrons transferred from the oxidized fatty acids during desaturation are transferred from the desaturases to cytochrome b5 and then NADH-cytochrome b5 reductase. These electrons are uncoupled from mitochondrial oxidative-phosphorylation and, therefore, do not yield ATP.
Since the mammalian desaturase enzymes cannot introduce sites of unsaturation beyond C9 they cannot synthesize either linoleic acid (18:2Δ9,12) or α-linolenic acid (ALA: 18:3Δ9,12,15). These two fatty acids must be acquired from the diet and are, therefore, referred to as essential fatty acids. Linoleic is especially important in that it required for the synthesis of arachidonic acid. Arachindonate is a precursor for the eicosanoids (the prostaglandins, thromboxanes, and leukotrienes). It is this role of fatty acids in eicosanoid synthesis that leads to poor growth, wound healing and dermatitis in persons on fat free diets. Also, linoleic acid is a constituent of epidermal cell sphingolipids that function as the skins’ water permeability barrier. The significance of ALA is that it serves as the precursor fatty acid for the synthesis of several clinically relevant omega-3 fatty acids, namely eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA).
The FADS3 encoded enzyme functions as a ceramide desaturase in the synthesis of the ceramide derivatives of the sphingadiene (SD) class. Sphingadienes are long chain sphingoid bases that contain an additional carbon-carbon double bond in the long-chain acyl moiety when compared to sphingosine. Sphingosine is the most common mammalian sphingoid base. FADS3 has been shown to be required for the synthesis of 4,14-sphingadiene (specifically: 4E,14Z-sphingadiene). The sphingadienes constitute a class of lipid that has been shown to exert cytotoxic effects on certain types of cancers through their ability to inhibit the activity of the AKT/PKB signaling pathway.
Clinical Significance of SCD-Mediated Desaturation
Stearoyl-CoA desaturase (SCD) is the rate-limiting enzyme catalyzing the synthesis of monounsaturated fatty acids (MUFAs). The expression of SCD is under the control of the transcription factor ChREBP as discussed above. The major products of SCD are oleic acid (18:1; a physiologically significant omega-9 fatty acid) and palmitoleic acid (16:1; a physiologically significant omega-7 fatty acid). These two monounsaturated fatty acids represent the majority of monounsaturated fatty acids present in membrane phospholipids, triglycerides, and cholesterol esters. Palmitoleic acid is the most abundant fatty acid in the blood and in adipose tissue. Palmitoleic acid possesses numerous biological activities and is referred to as a lipokine.
The ratio of saturated to monounsaturated fatty acids in membrane phospholipids is critical to normal cellular function and alterations in this ratio have been correlated with diabetes, obesity, cardiovascular disease, and cancer. Thus, regulation of the expression and activity of SCD has important physiological significance.
SCD activity has been shown to play a critical role in the prevention of cytotoxic stress. Cellular stress activates stress-activated kinases that in turn initiate a series of adaptive mechanisms that includes the unfolded protein response (UPR), autophagy, and apoptosis.
The effects of SCD in the regulation of cytotoxic stress are due to its role in the production of the MUFA oleic acid (18:1) and the subsequent generation of the phosphatidylinositol (PI) that contains oleic acid at both the sn1 and sn2 positions [1,2-dioleoyl-sn-glycero-3-phospho-(1’-myo-inositol)] designated PI[(18:1/18:1)]. This PI has been shown to inhibit p38 MAPK activation, counteract activation of the UPR and apoptosis, as well as regulating activation of autophagy. Given the role of PI[(18:1/18:1)] is maintenance of cellular integrity it has been define as a lipokine. The level of expression of the SCD gene has also been shown to be decreased during the onset of apoptosis and other forms of cell death. In addition, depletion of PI[(18:1/18:1)] has been linked to activation of cellular stress response pathways.
Synthesis and Biological Activities of Palmitoleic Acid
Palmitoleic acid can be acquired in the diet as well as synthesized from palmitic acid via the actions of SCD. Significant dietary sources of palmitoleic acid include salmon, cod liver oil, olive oil, chocolate, eggs, macadamia nut oil, and sea buckthorn.
The major site of synthesis of palmitic acid is adipose tissue. Another source of palmitoleic acid is hepatic synthesis of trans-palmitoleic acid from trans-vaccenic acid (trans-C18:1; an omega-7 MUFA) via peroxisomal β-oxidation. It was originally believed that trans-palmitoleic acid could be obtained only from the diet. This novel hepatic synthesis pathway represents a mechanism by which a dietary fatty acid (trans-vaccenic acid) can contribute to the accumulation of palmitoleic acid in numerous tissues.
Increased synthesis of palmitoleic acid is associated with enhanced peripheral insulin sensitivity and increased glucose uptake by the liver, adipose tissue, and skeletal muscle. Both the cis– and trans-palmitoleic acid forms are associated with enhanced insulin sensitivity and glucose homeostasis. Palmitoleic acid, produced by adipose tissue, acts in an autocrine manner to stimulate adipocyte lipolysis via increased PPARα activity resulting in increased adipose tissue triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) phosphorylation. Palmitoleic acid also stimulates insulin sensitivity of adipose tissue resulting in increased GLUT4 translocation to the plasma membranes of adipocytes and increased glucose uptake.
In mouse models of obesity and type 2 diabetes the administration of palmitoleic acid protects against hyperglycemia and hypertriglyceridemia, decreases the expression of hepatic lipogenic genes (SREBP1, FAS, and SCD), and results in reduced accumulation of triglycerides in the liver. Within the pancreas palmitoleic acid has been shown to stimulate pancreatic lipogenesis, phospholipid remodeling, and triglyceride synthesis. These effects of palmitoleic acid in the pancreas result in increased cell viability and endocrine functions of the organ.
Palmitoleic acid is an important regulator of cellular stress and inflammation, particularly in skeletal muscle and adipose tissue. Within skeletal muscle palmitoleic acid delays the activation of pro-oxidative and pro-inflammatory enzyme, COX2, and blocks the activation of p38 MAPK-mediated insulin resistance. Within adipocytes palmitoleic acid also exerts negative effects on the activation of inflammatory processes by inducing downregulation of genes encoding chemokines and cytokine receptors. Within the liver palmitoleic acid inhibits the expression of liver ER-stress genes and decreases expression of apoptotic genes. Palmitoleic acid also reduces apoptosis in pancreatic β-cells.
Exercise plays an important role in the level of circulating palmitoleic acid by activating adipose tissue lipolysis and release of stored palmitoleic acid. In addition, exercise increases de novo synthesis of palmitoleic acid by adipocytes. In mice in which the ATGL gene has been knocked out there is a failure of exercise-induced lipolysis, as well as in the normal process of cardiac hypertrophy and non-proliferative cardiomyocyte growth that normally occurs with exercise. When these mice are supplemented with palmitoleic acid there is restoration of left ventricular mass to the level of that in exercising wild-type mice. Exercise also leads to the accumulation of palmitoleic acid in triglycerides in skeletal muscle which has been shown to result in increased insulin sensitivity n skeletal muscle.
Clinical Significance of D5D and D6D Activities
Physiological and pathophysiological significance is also associated with the activities of D5D and D6D. The activity of D6D requires pyridoxal phosphate (PLP, derived from vitamin B6), Mg2+, and Zn2+ ions and its activity is, therefore, affected by nutritional status. Both of D5D and D6D exhibit reduced and inhibited activity in conditions associated with hyperglycemia such as is typical of type 2 diabetes. Additional factors impairing D5D and D6D activity include alcohol consumption, hypercholesterolemia, and the processes of aging. Conversely, normal insulin responsiveness results in increased D5D and D6D activity as does a caloric restriction diet.
Synthesis of Triglycerides
Fatty acids are stored for future use as triglycerides (TG) in all cells, but primarily in adipocytes of adipose tissue. TAGs constitute molecules of glycerol to which three fatty acids have been esterified. The fatty acids present in TAGs are predominantly saturated fatty acids. The fatty acids incorporated into TAGs are all activated to acyl-CoAs through the action of various acyl-CoA synthetases.
There are three major pathways for the synthesis of triglycerides referred to as the glycerol-3-phosphate (G3P) pathway, the dihydroxyacetone phosphate (DHAP) pathway, and the monoacylglycerol pathway. The glycerol-3-phosphate pathway of triglyceride synthesis is referred to as the Kennedy pathway after Eugene Kennedy who first worked out the pathway of triglyceride synthesis and published his results in 1960.
The glycerol-3-phosphate pathway is the major pathway (90%) utilized for triglyceride synthesis in hepatocytes. However, under conditions of insulin stimulation of the liver, glycolysis is highly active as a means to drive the excess glucose carbons into fatty acid synthesis. Under these conditions the DHAP pathway using the DHAP derived from glycolysis is the major triglyceride synthesis pathway in the liver. Within adipocytes, which lack expression of glycerol kinase, the DHAP derived from glycolysis is absolutely required for triglyceride synthesis thus, the DHAP pathway is the predominant triglyceride synthesis pathway in these cells. The monoacylglycerol pathway is used almost exclusively in enterocytes of the small intestines for the synthesis of triglycerides from dietary fatty acids.
Glyceroneogenesis and Triglyceride Synthesis
Although the DHAP derived from glycolysis is believed to be the primary source of the glycerol-3-phosphate used as the backbone for triglyceride synthesis, evidence clearly demonstrates that pyruvate (and by their relationship to pyruvate, alanine and lactate) can serve as the source of glycerol-3-phosphate via a process referred to as glyceroneogenesis.
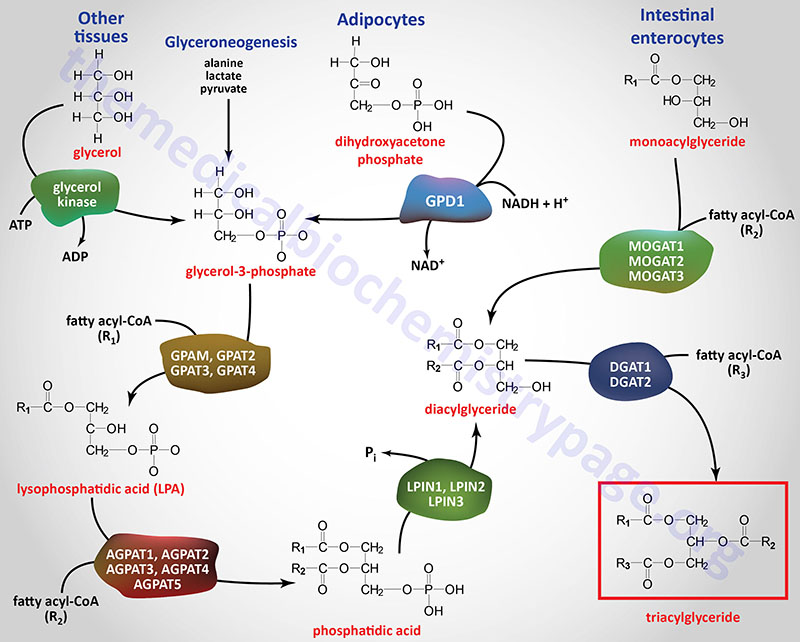
Glyceroneogenesis is essentially a diversion from the pathway of gluconeogenesis. Indeed, the pathways of gluconeogenesis and glyceroneogenesis in the liver share a common set of reactions, such that these pathways cannot be functionally separated. The cytosolic form of phosphoenolpyruvate carboxykinase (PEPCK-c: encoded by the PCK1 gene) catalyzes the GTP-dependent decarboxylation of oxaloacetate to form phosphoenolpyruvate which is important in the process of gluconeogenesis. The PCK1 encoded enzyme is a key regulatory enzyme in glyceroneogenesis.
In the process of glyceroneogenesis the carbons of pyruvate (or lactate or alanine) that are converted to glyceraldehyde-3-phosphate during gluconeogenesis are then preferentially diverted to DHAP as opposed to a 50% conversion as would be required in the pathway of gluconeogenesis. The DHAP is then reduced to glycerol-3-phosphate via the action of the cytoplasmic form of glycerol-3-phosphate dehydrogenase (encoded by the GPD1 gene) as opposed to condensation with glyceraldehyde-3-phosphate via the gluconeogenic action of aldolase A. The resulting glycerol-3-phosphate can then be used as the backbone for triglyceride synthesis.
Glycerol-3-Phosphate as Triglyceride Precursor
The glycerol backbone of TAG is activated by phosphorylation at the C-3 (sn-3) position by glycerol kinase generating glycerol-3-phosphate. Glycerol kinase is encoded by the GK gene. The GK gene is located on the X chromosome (Xp21.2) and is composed of 24 exons that generate four alternatively spliced mRNAs, each of which encode a distinct protein isoform.
Since expression of the GK gene is absent in adipocytes, these cells require the reduction of glycolytic DHAP to glycerol-3-phosphate via the action of cytosolic glycerol-3-phosphate dehydrogenase
The acylation of glycerol-3-phosphate is the major pathway for triglyceride synthesis in most human cells. Once formed, glycerol-3-phosphate is acylated at the C-1 (sn-1) position by one of a family of glycerol-3-phosphate acyltransferase (GPAT) enzymes that are expressed in either the mitochondria or in the endoplasmic reticulum, ER. The products of the acylation reactions are lysophosphatidic acids (LPA or lysoPA). Lysophosphatidic acids are themselves potent bioactive lipids. The LPA generated through the actions of the various GPAT enzymes serve, not only as substrates for triglyceride synthesis, but also for the synthesis of phospholipids.
The GPAT family includes the two mitochondrial enzymes (encoded by the GPAM and GPAT2 genes) and the two ER enzymes (encoded by the GPAT3 and GPAT4 genes). The GPAT3 and GPAT4 genes belong to the larger 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) family.
The GPAM gene is located on chromosome 10q25.2 and is composed of 26 exons that generate two alternatively spliced mRNAs encoding the same 828 amino acid precursor protein. GPAM is also known as GPAT1. The highest levels of expression of the GPAM gene are seen in liver and adipose tissues. Expression of the GPAM gene decreases during fasting and increases following food intake, the latter the result of effects of insulin. GPAM exhibits a preference for palmitic acid (C16:0).
The GPAT2 gene is located on chromosome 2q11.1 and is composed of 23 exons that generate nine alternatively spliced mRNAs, that collectively encode seven distinct protein isoforms. Expression of the GPAT2 gene is nearly exclusive to the germ cells of the testes. Substrate preference for GPAT2 appears to be for arachidonic acid (C20:4).
The GPAT3 gene is located on chromosome 4q21.23 and is composed of 17 exons that generate three alternatively spliced mRNAs all of which encode the same 434 amino acid protein. Confusion results from the fact that GPAT3 has also been identified as AGPAT8, AGPAT9, and AGPAT10 where the latter two designations have also been associated with LPCAT1. Expression of GPAT3 increases during adipogenesis indicating that the encoded enzyme plays a central role in triglyceride storage in adipose tissue.
The GPAT4 gene is located on chromosome 8p11.21 and is composed of 17 exons that generate three alternatively spliced mRNAs that collectively encode two distinct protein isoforms. Isoform 1 is 456 amino acids and isoform 2 is 248 amino acids. GPAT4 is also known as AGPAT6 and lysophosphatidic acid acyltransferase-zeta (LPAATζ). Expression of the GPAT4 gene is highest in the liver and in white and brown adipose tissue, WAT and BAT, respectively. GPAT4 exhibits a preference of monounsaturated fatty acids (MUFA).
All of the GPAT enzymes are members of the large family of 1-acylglycerol-3-phosphate O-acyltransferases (AGPAT) as outlined in the following section.
Acylglycerophosphate acyltransferases: AGPAT
Following formation of a lysophosphatidic acid, various 1-acylglycerol-3-phosphate O-acyltransferases (AGPAT) add another CoA-activated fatty acid to the sn-2 position generating 1,2-diacylglycerol phosphates which are commonly identified as phosphatidic acids (PA). Several enzymes of the AGPAT family are also referred to as lysophospholipid acyltransferases (LPLAT) as well as lysophosphatidic acid acyltransferases (LPAAT). Several enzymes in the LPLAT family are involved in the synthesis of the glycerophopholipids from lysophosphatidic acids and from lysophospholipids as discussed in the Phospholipid Synthesis section below.
There are five major 1-acylglycerol-3-phosphate O-acyltransferase encoding genes identified as AGPAT1–AGPAT5 that function in the synthesis of triglycerides. All of these AGPAT enzymes are integral membrane proteins with the AGPAT1 and AGPAT2 enzymes specifically localized to the ER.
The AGPAT1 gene is located on chromosome 6p21.32 and is composed of 11 exons that generate five alternatively spliced mRNAs, four of which encode the same 283 amino acid protein. The AGPAT1 encoded enzyme is also known as LPLAT1 and as LPAATα (alpha). The AGPAT1 gene is ubiquitously expressed. The AGPAT1 enzyme is predominantly localized to the endoplasmic reticulum, ER.
The AGPAT2 gene is located on chromosome 9q34.3 and is composed of 7 exons that generate two alternatively spliced mRNAs encoding two different isoforms of the enzyme. The AGPAT2 encoded enzyme is also known as LPLAT2 as well as LPAATβ (beta). The AGPAT2 enzyme is predominantly localized to the endoplasmic reticulum, ER. Mutations in the AGPAT2 gene result in congenital generalized lipodystrophy 1 (CGL1), also known as Berardinelli-Seip syndrome. This disorder is an autosomal recessive disorder characterized by marked lack of adipose tissue at birth, severe insulin resistance, hypertriglyceridemia, hepatic steatosis, and early onset of diabetes.
The AGPAT3 gene is located on chromosome 21q22.3 and is composed of 22 exons that generate five alternatively spliced mRNAs, four of which encode the same 376 amino acid protein. The AGPAT3 encoded enzyme is also known LPLAT3 as well as LPAATγ (gamma). The AGPAT3 gene is ubiquitously expressed with highest levels in heart, adipose tissue, and liver. The AGPAT3 enzyme is predominantly localized to the endoplasmic reticulum (ER) where it is involved in retrograde transport processes from the Golgi back to the ER.
The AGPAT4 gene is located on chromosome 6q26 and is composed of 9 exons that encode a 378 amino acid protein. The AGPAT4 encoded enzyme is also known as LPLAT4 as well as LPAATδ (delta). The AGPAT4 gene is predominantly expressed in the brain and muscle. The encoded protein localizes to the outer membrane of the mitochondria and to the trans-Golgi network.
The AGPAT5 gene is located on chromosome 8p23.1 and is composed of 9 exons that encode a 364 amino acid protein. The AGPAT5 encoded enzyme is also known as LPLAT5 as well as LPAATε (epsilon).
Humans express six additional genes in the large AGPAT family, two of which are the GPAT3 (also known as AGPAT9) and GPAT4 (also known as AGPAT6) genes indicated above. Four of the AGPAT family member genes encode enzymes involved in the synthesis of glycerophospholipids (phospholipids).
The 11 genes that constitute the mammalian AGPAT family are categorized into three groups identified as group I, II, and II. Group III is further divided into three subgroups, IIIa, IIIb, and IIIc. Group I genes include AGPAT1 and AGPAT2. Group II genes include GPAT3 (AGPAT9), GPAT4 (AGPAT6), LPCAT1 (AGPAT10), LPCAT2 (AGPAT11), and LPCAT4 (AGPAT7). Group IIIa genes include AGPAT3 and AGPAT4. Group IIIb comprises the LCLAT1 (AGPAT8) gene. Group IIIc comprises the AGPAT5 gene.
Monoacylglycerides as Triglyceride Precursors
Intestinal monoacylglycerides (MAG), derived from the hydrolysis of dietary fats, can also serve as substrates for the synthesis of triglycerides (TAG) which can then be incorporated into chylomicrons. The intestinal synthesis of a TAG from a MAG first involves MAG acylation carried out by one of a family of monoacylglycerol O-acyltransferases (MOGAT). Humans express three MOGAT genes identified as MOGAT1, MOGAT2, and MOGAT3.
The MOGAT1 gene is located on chromosome 2q36.1 and is composed of 6 exons that encode a 335 amino acid protein.
The MOGAT2 gene is located on chromosome 11q13.5 and is composed of 8 exons that encode a 334 amino acid protein.
The MOGAT3 gene is located on chromosome 7q22.1 and is composed of 8 exons that generate two alternatively spliced mRNAs encoding two distinct isoforms of the enzyme.
The resultant DAG, in intestinal enterocytes, are then converted to TAG via the action of the DGAT enzymes outlined in the next paragraph.
The phosphate of phosphatidic acid is removed, by phosphatidic acid phosphatase (PAP1), to yield 1,2-diacylglycerols (DAG). The PAP1 enzyme is encoded by a gene that is a member of the lipin family (see next section) identified as the LPIN1 gene. The DAG, along with a fatty acyl-CoA, serve as substrates for a diacylglycerol O-acyltransferase (DGAT) enzyme forming a triglyceride, TAG. Humans express two DGAT encoding genes identified as DGAT1 and DGAT2.
The DGAT1 gene is located on chromosome 8q24.3 and is composed of 21 exons that encode a 488 amino acid protein.
The DGAT2 gene is located on chromosome 11q13.5 and is composed of 10 exons that generate two alternatively spliced mRNAs encoding two isoforms of the enzyme.
Glycerolipid-Fatty Acid Cycle
The glycerolipid-fatty acid cycle represents a central pathway of overall energy homeostasis. Th cycle integrates glucose and lipid metabolism and also generates bioactive lipid molecules, the lysophosphatidic acids (LPA or LysoPA). When fuels are abundant, excess glucose and free fatty acids are diverted into triglyceride and glycerolipid synthesis. The glucose is metabolized to glycerol-3-phosphate and the fatty acids are esterified to this molecule to generate triglycerides. However, in the context of the glycerolipid-fatty acid cycle the synthesis of lysoPA, phosphatidic acid (PA), and phospholipids also takes place.
The glycerolipid-fatty acid cycle consists of both lipid synthesis (lipogenesis) and lipid metabolism (lipolysis) segments and as such serves to generate complex bioactive lipids that control many biological processes. These biological processes include, but are not limited to, the synthesis, secretion, and function of insulin. Indeed, the glycerolipid-fatty acid cycle is disturbed in obesity and type 2 diabetes.
During lipogenesis reactions the glycerol-3-phopshate derived from glycolysis is esterified, as described above, to generate triglycerides. As indicate, during triglyceride synthesis the intermediates, lysoPA, PA, and diacylglycerides all constitute bioactive lipids. When triglycerides are broke down, in the context of lipolysis reactions, diacylglycerides, monoacylglycerides, free fatty acids, and glycerol are generated.
The function of the glycerolipid-fatty acid cycle can be regulated through the dephosphorylation of glycerol-3-phosphate to glycerol. This dephosphorylation is catalyzed by an enzyme identified as phosphoglycolate phopshatase. Phosphoglycolate phosphatase is encoded by the PGP gene. The PGP gene is located on chromosome 16p13.3 and is composed of 2 exons that encode a 321 amino acid protein. Expression of the PGP gene is ubiquitous with highest level seen in testes, heart, skeletal muscle, and pancreas. The high level of expression of PGP in heart and skeletal muscle is likely to ensure that toxic levels of lipids do not accumulate in these two tissues that prefer to oxidize fatty acids for energy production.
The level of phosphoglycolate phosphatase activity is directly correlated to the control of glycolysis, lipogenesis, lipolysis, fatty acid oxidation, mitochondrial energy metabolism, and the cellular redox state, particularly in pancreatic β-cells and hepatocytes. In addition, the activity of phosphoglycolate phosphatase is involved in the regulation of glucose-stimulated insulin secretion (GSIS), the response of pancreatic β-cells to stress, and in control of gluconeogenesis in hepatocytes.
Lipin Genes: TAG Synthesis and Transcriptional Regulation
In addition to its role in overall TAG and phospholipid homeostasis, phosphatidic acid phosphatase (PAP1: encoded by the LPIN1 gene) has been shown to possess additional important activities. The lipin-1 encoding gene (LPIN1) was originally identified as being mutated in a line of mice possessing a fatty liver dystrophy syndrome. This mouse line is identified as the fld mouse.
Subsequent to the identification of the LPIN1 gene in the fld mouse, two additional related genes were identified. These three lipin genes are identified as LPIN1, LPIN2, and LPIN3. All characterized lipin proteins possess phosphatidic acid phosphatase activity that is dependent upon Mg2+ or Mn2+ and phosphatidic acid as the substrate.
The LPIN1 gene is located on chromosome 2p25.1 and is composed of 29 exons that generate 13 alternatively spliced mRNAs that collectively encode nine distinct protein isoforms.
The LPIN2 gene is located on chromosome 18p11.31 and is composed of 25 exons that generate three alternatively spliced mRNAs, each of which encode the same 896 amino acid protein. Mutations in the LPIN2 gene are associated with Majeed syndrome which is characterized by chronic recurrent osteomyelitis, cutaneous inflammation, recurrent fever, and congenital dyserythropoietic anemia.
The LPIN3 gene is located on chromosome 20q12 and is composed of 24 exons that generate two alternatively spliced mRNAs, both of which encode distinct protein isoforms.
In addition to the obvious role of lipin-1 in TAG synthesis, evidence indicates that the protein is also required for the development of mature adipocytes, coordination of peripheral tissue glucose and fatty acid storage and utilization, and serves as a transcriptional co-activator. The latter function has significance to diabetes as it has been shown that some of the effects of the thiazolidinedione (TZD) class of drugs used to treat the hyperglycemia associated with type 2 diabetes are exerted via the effects of lipin-1. Although the lipin proteins do not contain DNA-binding motifs they have protein-interaction domains that allow them to form complexes with nuclear receptors and function as transcriptional regulators.
Lipin-1 has been shown to interact with peroxisome proliferator-activated receptor-γ [PPARγ] co-activator 1α (PGC-1α) and PPARα leading to enhanced gene expression. Lipin-1 also is known to interact with additional members of the nuclear receptor family including the glucocorticoid receptor (GR) and hepatocyte nuclear factor-4α (HNF-4α).
Lipin-1 also induces the expression of the adipogenic transcription factors PPARγ and CCAAT-enhancer-binding protein α (C/EBPα). The functions of lipin-1α and lipin-1β appear to be complimentary with respect to adipocyte differentiation. Lipin-1α induces genes that promote adipocyte differentiation while lipin-1β induces the expression of lipid synthesizing genes such as fatty acid synthase (FAS) and diacylglycerol acyltransferase (DGAT). The interactions of lipin-1 with PPARα and PGC-1α leads to increased expression of fatty acid oxidizing genes such as carnitine palmitoyltransferase 1 (CPT1: encoded by the CPT1A gene), acyl-CoA oxidases (peroxisomal fatty acid oxidation pathway), and medium-chain acyl-CoA dehydrogenase (MCAD: encoded by the ACADM gene).
Phospholipid Structures
Phospholipids are synthesized by esterification of an alcohol to the phosphate of phosphatidic acid (1,2-diacylglycerol 3-phosphate) or through the remodeling of lysophospholipids. Most phospholipids have a saturated fatty acid on C-1 (sn-1) and an unsaturated fatty acid on C-2 (sn-2) of the glycerol backbone. In humans, the fatty acids esterified to phosphatidic acid are most often 12 to 24 carbons in length and contain from 0 to 6 carbon-carbon double bonds. The most commonly added alcohols (serine, ethanolamine and choline) also contain nitrogen that may be positively charged, whereas, glycerol and inositol do not.
The major classifications of phospholipids are the phosphatidylserines (PS), the phosphatidylethanolamines (PE), the phosphatidylcholines (PC; often referred to as lecithins), the phosphatidylinositols (PI), phosphatidylglycerols (PG; major components of pulmonary surfactant), and the diphosphatidylglycerols (DPG; more commonly called the cardiolipins).
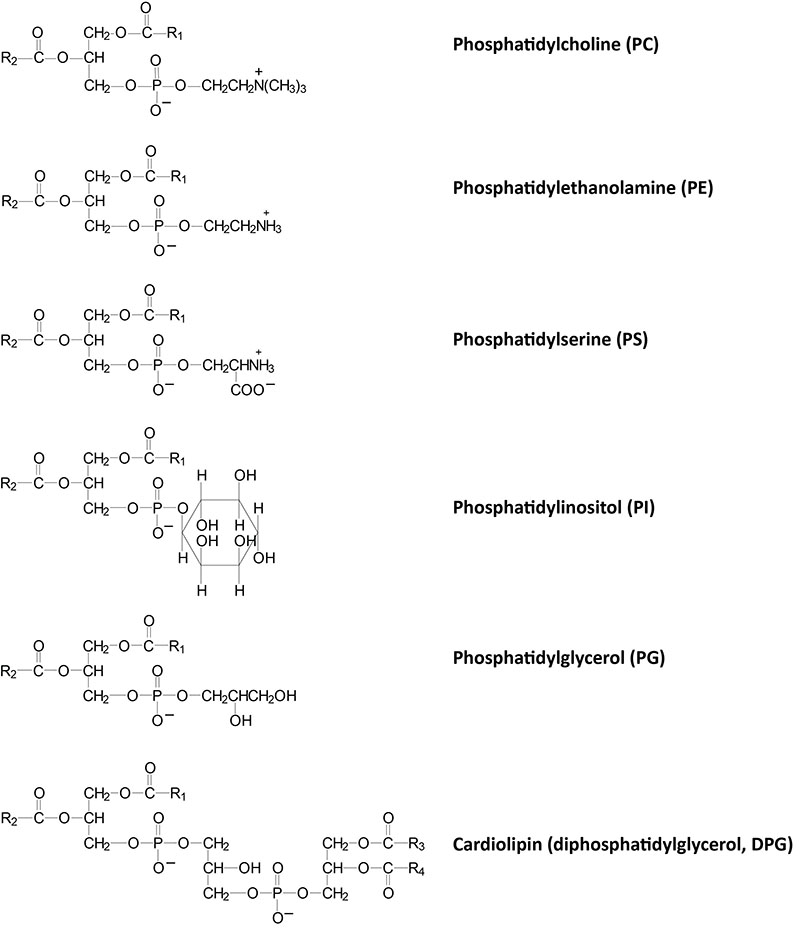
Table of Phospholipid Composition of Various Mammalian Membranes (% total phospholipid)
| Membrane Type | ||||||
| Phospholipid Class | Plasma | Mitochondrial | Nuclear | ER | Golgi | Lysosomal |
| Phosphatidylserine | 4% | 1% inner; 1% outer | 6% | 4% | 4% | 1% |
| Phosphatidylethanolamine | 21% | 38% inner; 34% outer | 25% | 21% | 17% | 21% |
| Phosphatidylcholine | 43% | 41% inner; 49% outer | 52% | 57% | 4% | 42% |
| Phosphatidylinositol | 4% | 2% inner; 9% outer | 4% | 9% | 9% | 6% |
| Cardiolipin | 0% | 16% inner; 5% outer | 0% | 0% | 0% | 0% |
Phospholipid Synthesis
Synthesis of the phospholipids occurs via a de novo pathway referred to as the Kennedy pathway. This pathway involves the fatty acid esterification of glycerol-3-phosphate at the sn-1 and sn-2 positions generating a phosphatidic acid followed by addition of the polar head group. The action of phospholipases generates lysophospholipids which can be re-esterified via the action of various enzymes of the lysophospholipid acyltransferase (LPLAT) family resulting in remodeling of phospholipids. This latter pathway to phospholipid synthesis is termed the Lands pathway.
Enzymes of the LPLAT family belong to two subfamilies of functionally related enzymes identified as the AGPAT family and the membrane bound O-acyltransferase domain containing (MBOAT) family. As discussed above in the Acylglycerophosphate acyltransferases: AGPAT section, several AGPAT family enzymes are involved in the synthesis of triglycerides. The AGPAT family consists of 11 genes in humans and the MBOAT family consists of 8 genes in humans. Not all of these 19 genes encode enzymes involved in phospholipid synthesis. For example, the HHAT encoded hedgehog acyltransferase is an MBOAT family enzyme and it is responsible for the acylation of sonic hedgehog.
Table of the Human Lysophospholipid Acyltransferase Family
| Official Gene Name | LysoPhosphoLipid AcylTransferase (LPLAT) Name | Other Names | Primary Tissues | Functions / Comments |
| AGPAT1 | LPLAT1 | LPAATα, LPAAT1 | ubiquitously expressed | localized to the endoplasmic reticulum (ER); involved in triglyceride synthesis in addition to phospholipid synthesis; plays a crucial role in the central nervous system and the reproductive system; polymorphisms in AGPAT1 gene associated with exfoliation syndrome |
| AGPAT2 | LPLAT2 | LPAATβ, LPAAT2 | highest in adipose tissue; 4-5-fold lower levels in kidney, liver, lung, intestines, skeletal muscle | localized to the endoplasmic reticulum (ER); involved in triglyceride synthesis in addition to phospholipid synthesis; play a crucial role in triglyceride production in adipose tissues; mutations in AGPAT2 gene associated with type 1 congenital generalized lipodystrophy (CGL) which is also known as Berardinelli–Seip lipodystrophy |
| AGPAT3 | LPLAT3 | LPAATγ, LPAAT3 | highest levels in kidney with brain being next highest; ubiquitously expressed | localized to the endoplasmic reticulum (ER), Golgi, and nuclear envelope; involved in triglyceride synthesis in addition to phospholipid synthesis; primarily utilizes docosahexaenoic acid (DHA, C22:6) for phospholipid synthesis |
| AGPAT4 | LPLAT4 | LPAATδ, LPAAT4 | highest levels in brain, 4-fold lower levels in several tissues | localized to the endoplasmic reticulum (ER); involved in triglyceride synthesis in addition to phospholipid synthesis; primarily utilizes unsaturated fatty acids as substrate including docosahexaenoic acid (DHA, C22:6), arachidonic acid (C20:4), linoleic acid (C18:2), and oleic acid (C18:1) |
| AGPAT5 | LPLAT5 | LPAATε, LPAAT5 | ubiquitously expressed with highest levels in brain and testis | localized to the endoplasmic reticulum (ER); involved in triglyceride synthesis in addition to phospholipid synthesis; preference for oleic acid (C18:1); polymorphisms associated with insulin resistance; experimental reductions in levels of enzyme shown to improve insulin sensitivity |
| LCLAT1 | LPLAT6 | AGPAT8, ALCAT1 | ubiquitously expressed with highest levels in the intestines | localized to the endoplasmic reticulum (ER); acylates numerous lysoPL substrates; exhibits preference for stearic acid (C18:0) and lysophosphatidylinositol (lysoPI); also utilizes lysophosphatidylglycerol (lysoPG) and lysocardiolipin (lysoCL) as substrates; is involved in oxidative stress and mitochondrial dysfunction via its role in cardiolipin remodeling |
| LPCAT1 | LPLAT8 | AGPAT9, AGPAT10 | lung, spleen | localized to the endoplasmic reticulum (ER) and lipid droplets (LD); primarily involved in the synthesis of phosphatidylcholines (PC); utilizes palmitic acid (C16:0) as a substrate; is involved in the synthesis of dipalmitoylphosphatidylcholine (DPPC; also termed dipalmitoyllecithin) which is a critical component of pulmonary surfactant; function of LPCAT1 may contribute to progression of numerous cancers |
| LPCAT2 | LPLAT9 | AGPAT11, LysoPAFAT | thyroid gland; 5-fold lower levels in many other tissues | localized to the endoplasmic reticulum (ER) and lipid droplets (LD); primarily responsible for the synthesis of platelet activating factor (PAF); utilizes acetic acid for incorporation into lyso-PAF; activity is dependent on Ca2+; only enzyme of this family that is regulated by phosphorylation |
| LPCAT3 | LPLAT12 | MBOAT5 | ubiquitously expressed with highest levels in the intestines with high levels also in liver | localized to the endoplasmic reticulum (ER); preference for linoleic acid (C18:2) and arachidonic acid (C20:4) when acylating lysophosphatidylcholine (lysoPC), lysophosphatidylethanolamine (lysoPE), and lysophosphatidylserine (lysoPS); plays an essential role in lipid mobilization in the gut and the liver; plays a role in the regulation of gut-brain communication through regulated gut hormone secretion; loss of LPCAT3 function is associated with lipid malabsorption |
| LPCAT4 | LPLAT10 | AGPAT7, LPEAT2 | ubiquitously expressed with highest levels in the stomach | localized to the endoplasmic reticulum (ER); exhibits broad substrate specificity and utilizes palmitic acid (C16:0), stearic acid (C18:0), and oleic acid (C18:1) in the production of phosphatidylethanolamines (PE), phosphatidylcholines (PC), and phosphatidylglycerols (PG); also acylates alkyl-lysoPC and alkenyl-lysoPE; may also utilize DHA as a substrate |
| LPGAT1 | LPLAT7 | FAM34A | ubiquitously expressed with highest levels in the small intestines and thyroid gland | localized to the endoplasmic reticulum (ER); primarily acylates lysophosphatidylglycerols (lysoPG) but is involved in the synthesis of numerous phospholipids; utilizes palmitic acid (C16:0), stearic acid (C18:0), and oleic acid (C18:1); polymorphisms in gene associated with susceptibility to obesity in Pima Indians |
| MBOAT1 | LPLAT14 | OACT1, LPEAT1 | ubiquitously expressed with highest levels in the stomach and intestines | localized to the endoplasmic reticulum (ER); preference for oleic acid (C18:1) but also utilizes palmitic acid (C16:0) and arachidonic acid (C20:4) |
| MBOAT2 | LPLAT13 | OACT2, LPEAT, LPAAT | ubiquitously expressed with highest levels in the brain and bone marrow | localized to the endoplasmic reticulum (ER); exhibits selectivity for incorporation of oleic acid (C18:1) into lysophosphatidylcholine (lysoPC) and lysophosphatidylethanolamine (lysoPE) |
| MBOAT7 | LPLAT11 | LENG4, LPIAT1 | ubiquitously expressed with highest levels in the adrenal gland, brain, and testis | localized to the endoplasmic reticulum (ER); LPIAT1 is lysophosphatidylinositol acyltransferase 1; exhibits selectivity for incorporation of arachidonic acid (C20:4) into lysophosphatidylinositol (lysoPI); loss of function is associated with abnormal brain development; polymorphisms in gene associated with hepatic steatosis |
Phosphatidylcholines, PC
This class of phospholipid is also called the lecithins. The phosphatidylcholines represent the predominant phospholipid in the membranes of human cells, representing >50% of the phospholipid composition of membranes. At physiological pH, phosphatidylcholines are neutral zwitterions. They contain primarily palmitic or stearic acid at carbon 1 and primarily oleic acid (18:1), linoleic acid (18:2), or linolenic acid (18:3) at carbon 2. The specific phosphatidylcholine with both C-1 and C-2 esterified with palmitic acid (16:0) is commonly identified as dipalmitoyllecithin. Dipalmitoyllecithin is a component of pulmonary surfactant and is the major (80%) phospholipid found in the extracellular lipid layer lining the pulmonary alveoli.
Phosphatidylcholine Synthesis and Respiratory Distress Syndrome (RDS)
A significant cause of death in premature infants and, on occasion, in full term infants is respiratory distress syndrome (RDS) or hyaline membrane disease. This condition is caused by an insufficient amount of pulmonary surfactant. Under normal conditions the surfactant is synthesized by type II endothelial cells and is secreted into the alveolar spaces to prevent atelectasis following expiration during breathing. Surfactant is comprised primarily of dipalmitoyllecithin. Additional lipid components include phosphatidylglycerol and phosphatidylinositol. In addition to the lipid composition surfactant contains proteins of 18 kDa and 36 kDa (termed surfactant proteins). During the third trimester the fetal lung synthesizes primarily sphingomyelin, and type II endothelial cells convert the majority of their stored glycogen to fatty acids and then to dipalmitoyllecithin. Fetal lung maturity can be determined by measuring the ratio of lecithin to sphingomyelin (L/S ratio) in the amniotic fluid. An L/S ratio less than 2.0 indicates a potential risk of RDS. The risk is nearly 75-80% when the L/S ratio is 1.5.
Phosphatidylcholine Synthesis
In humans, phosphatidylcholines are primarily synthesized via the CDP-choline pathway (Kennedy pathway). The choline required for the synthesis of phosphatidylcholines is primarily acquired in the diet. Indeed, one of the major fates of dietary choline is the incorporation into phosphatidylcholines.
Dietary choline, as well as that recycled from phosphatidylcholines, is converted to betaine (trimethylglycine) which plays an important role in in the synthesis of methionine from homocysteine and as a consequence, contributes to numerous methylation reaction when methionine is converted to S-adenosylmethionine (SAM or AdoMet).
Choline is first activated by phosphorylation and then by coupling to CDP prior to attachment to a 1,2-diacylglycerol. The phosphorylation of choline is catalyzed by choline kinase-α (encoded by the CHKA gene). Humans express two choline kinase genes, CHKA and CHKB (choline kinase-β; formerly called choline kinase-like, CHKL). The CHKA encoded enzyme is also responsible for the phosphorylation and activation of ethanolamine.
Phosphocholine is then converted to CDP-choline by the enzymes of the cytidylyltransferase family. Humans express three genes in this family, PCYT1A (phosphate cytidylyltransferase 1, choline, alpha), PCYT1B (phosphate cytidylyltransferase 1, choline, beta), and PCYT2 (phosphate cytidylyltransferase 2, ethanolamine). The PCYT1A and PCYT1B encoded enzymes are involved in CDP-choline synthesis while the PCYT2 encoded enzyme is involved in CDP-ethanolamine synthesis. The PCYT1A enzyme contains a nuclear localization signal and thus, it is predominantly found in this compartment.
Phosphatidylcholines are then synthesized from CDP-choline and a 1,2-diacylglycerol with concomitant release of CMP. The 1,2-diacylglycerols are derived from the action of enzymes of the phospholipid phosphatase family, such as lipin-1 (encoded by the LPIN1 gene), phospholipid phosphatase 1 (encoded by the PLPP1 gene), phospholipid phosphatase 2 (encoded by the PLPP2 gene), and phospholipid phosphatase 3 (encoded by the PLPP3 gene). The last reaction of PC synthesis is catalyzed by diacylglycerol cholinephosphotransferase which is encoded by the CHPT1 (choline phosphotransferase 1) gene. The CHPT1 encoded enzyme is often referred to as CDP-choline:1,2-diacylglycerol cholinephosphotransferase.
An additional pathway for the synthesis of PC involves the trimethylation of PE using S-adenosylmethionine (SAM; or AdoMet) as methyl group donor. This second PC synthesis pathway only occurs to a significant degree in hepatocytes and is catalyzed by phosphatidylethanolamine N-methyltransferase which is encoded by the PEMT gene. Although an additional PC biosynthesis pathway is known that involves the addition of choline to CDP-activated 1,2-diacylglycerol, this pathway is not known to occur in human cells..
Phosphatidylethanolamines, PE
The phosphatidylethanolamines are neutral zwitterions at physiological pH. They contain primarily palmitic or stearic acid on carbon 1 and a long chain unsaturated fatty acid (e.g. 18:2, 20:4 and 22:6) on carbon 2. Synthesis of PE in humans is carried out by at least two distinct pathways and they account for the bulk of PE synthesis . These two pathways operate either in the ER or in the mitochondria.
One of the two major PE synthesis pathway occurs in the ER and involves the phosphorylation of ethanolamine via the action of CHKA encoded enzyme described in the section above. Phosphoethanolamine is then converted to CDP-ethanolamine via the activity of the PCYT2 encoded enzyme described in the previous section. Formation of PE from CDP-ethanolamine and a 1,2-diacylglycerol is catalyzed by selenoprotein I which is encoded by the SELENOI gene. The encoded enzyme is more commonly referred to as CDP-ethanolamine:1,2-diacylglycerol ethanolamine phosphotransferase and also as diacylglycerol ethanolamine phosphotransferase. The second major pathway for PE synthesis occurs in the inner mitochondrial membrane and involves the decarboxylation of PS. The decarboxylation reaction is catalyzed by phosphatidylserine decarboxylase which is encoded by the PISD gene.
Another minor PE synthesis reaction involves the fatty acylation of a lysophosphatidylethanolamine (lyso-PE) converting it to PE which is catalyzed by an enzyme called lyso-PE acyltransferase which is encoded by the MBOAT2 (membrane bound O-acyltransferase domain containing 2) gene.
Phosphatidylserines, PS
Phosphatidylserines will carry a net charge of –1 at physiological pH and are composed of fatty acids similar to the phosphatidylethanolamines. The pathway for PS synthesis involves base exchange reactions of serine for the ethanolamine in PE or serine for the choline in PC. These exchange reactions can be catalyzed by a single enzyme, phosphatidylserine synthase 1, which is encoded by the PTDSS1 gene. The enzyme encoded by the PTDSS1 gene exhibits a high degree of preference for PC and is, therefore, the primary enzyme for generation of PS via the base exchange reaction. A related gene, PTDSS2, encodes phosphatidylserine synthase 2 and this enzymes appears to be exclusive for the exchange of serine for ethanolamine in PE generating PS. Also, as indicated above, PS can serve as a source of PE through a decarboxylation reaction catalyzed by the PISD encoded enzyme.
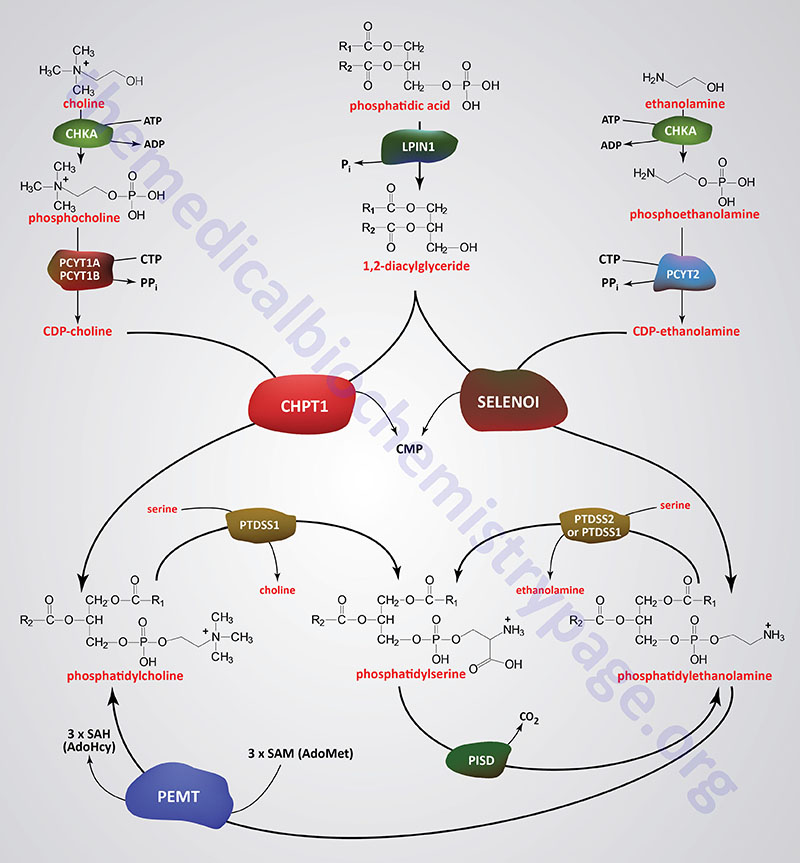
Phosphatidylinositols (PI or PtdIns)
The phosphatidylinositols contain almost exclusively stearic acid at carbon 1 and arachidonic acid at carbon 2. Phosphatidylinositols composed exclusively of non-phosphorylated inositol exhibit a net charge of –1 at physiological pH. These molecules exist in membranes with various levels of phosphate esterified to the hydroxyls of the inositol. Molecules with phosphorylated inositol are termed phosphoinositides. The phosphoinositides are important intracellular transducers of signals emanating from the plasma membrane.
In humans, the synthesis of a PI (as well as a PG) begins with the formation of CDP-diacylglycerol from phosphatidic acid and CTP. The synthesis of CDP-diacylglycerol is catalyzed by at least three enzymes that includes CDP-diacylglycerol synthase 1 and CDP-diacylglycerol synthase 2, which are encoded by the CDS1 and CDS2 genes, respectively. The third enzyme that can synthesize CDP-diacylglycerol is a bifunctional enzyme that is also responsible for the condensation of a CDP-diacylglycerol with myo-inositol forming a PI. This latter enzyme is called CDP-diacylglycerol–inositol 3-phosphatidyltransferase and it is encoded by the CDIPT gene. The CDIPT encoded enzyme is also referred to simply as phosphatidylinositol synthase, PIS. In humans the major enzyme responsible for the synthesis of CDP-diacylglycerol used for PI synthesis is likely to be the CDS1 encoded enzyme.
Phosphatidylinositols subsequently undergo a series of phosphorylations of the hydroxyls of inositol leading to the production of polyphosphoinositides. One particular polyphosphoinositide, phosphatidylinositol (4,5)-bisphosphate (PIP2; also designated PtdIns-4,5-P2), is a critically important membrane phospholipid involved in the transmission of signals for cell growth and differentiation.
Another critically important PI is phosphatidylinositol (3,4,5)-trisphosphate (PIP3; also designated PtdIns-3,4,5-P3) which is generated from PIP2 via the action of the phosphatidylinositol-3-kinases, PI3K. PIP3 activates the kinase called phosphoinositide-dependent kinase-1 (PDK1) which serves as a master regulatory kinase in the activation of the kinases PKB/AKT, calcium and phospholipid-dependent kinase, PKC, S6K (p70-S6 kinase 1), and the Ser/Thr kinases identified as SGK1, SGK2, and SGK3. PKB/AKT was originally identified as the tumor inducing gene in the AKT8 retrovirus found in the AKR strain of mice. Humans express three genes in the AKT family identified as AKT1 (PKBα), AKT2 (PKBβ), and AKT3 (PKBγ).
Phosphatidylglycerols, PG
Phosphatidylglycerols exhibit a net charge of –1 at physiological pH. These molecules are found in high concentration in mitochondrial membranes and as components of pulmonary surfactant. Phosphatidylglycerols are synthesized in a two-step process that begins with CDP-diacylglycerol and glycerol-3-phosphate. The first reaction yields phosphatidylglycerol phosphate and this reaction is catalyzed by the ER-localized enzyme identified as phosphatidylglycerophosphate synthase 1 (encoded by the PGS1 gene). The CDP-diacylglycerol is synthesized by the CDS1, CDS2, or CDIPT encoded enzymes as described in the previous section.
Phosphatidylglycerol phosphates are then converted to phosphatidylglycerols (PG) via removal of phosphate most likely by the action of the PGS1 encoded enzyme although the activity has been called PGP phosphatase, phosphatidylglycerol phosphate phosphatase, and phosphatidylglycerophosphatase. Phosphatidylglycerols are also the precursors for the synthesis of the diphosphatidylglycerols (DPG) which are more commonly called the cardiolipins. The cardiolipins are major lipid components of the inner mitochondrial membrane.
Diphosphatidylglycerols: Cardiolipins
The cardiolipins are very acidic, exhibiting a net charge of –2 at physiological pH. They are found almost exclusively in the inner mitochondrial membrane. The cardiolipins are synthesized by the condensation of a CDP-diacylglycerol with a PG in a reaction catalyzed by cardiolipin synthase 1 which is encoded by the CRLS1 gene.
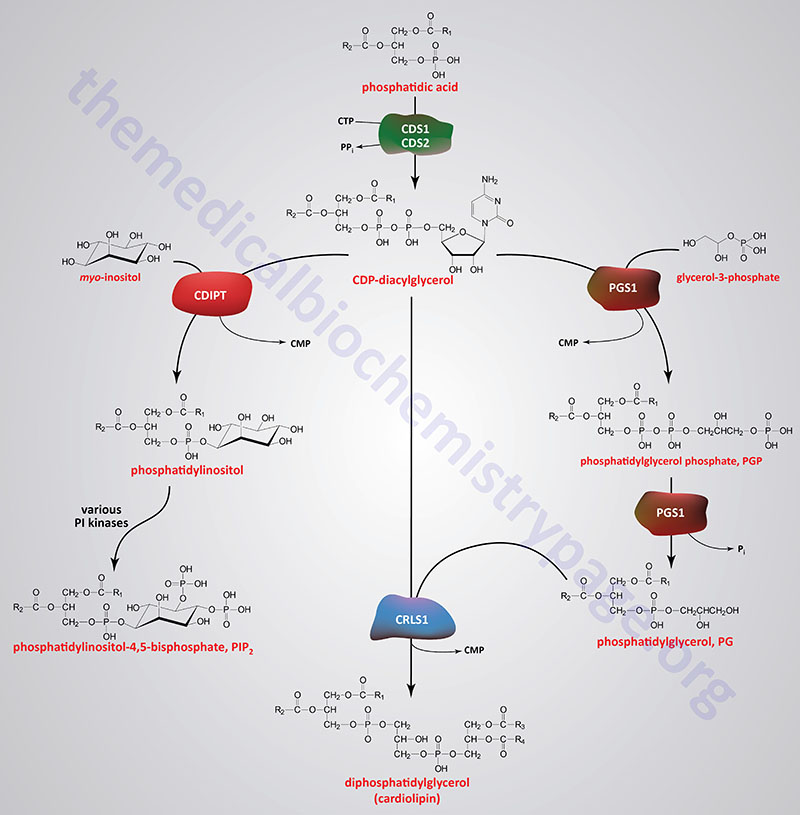
Acylglycerol Kinase and Mitochondrial Phospholipids
Acylglycerol kinase is a phospholipid kinase that is primarily localized to the mitochondrial inner membrane where it functions in mitochondrial phospholipid homeostasis. Acylglycerol kinase is also identified as multi-substrate lipid kinase (MULK). The identification of AGK came about through an investigation of the underlying cause of Sengers syndrome. Sengers syndrome is a disorder that is characterized by hypertrophic cardiomyopathy, cataracts, and lactic acidosis, with or without skeletal myopathy.
Acylglycerol kinase catalyzes the synthesis of mitochondrial lysophosphatidic acid (LPA) and phosphatidic acid (PA) from monoacylglycerides (MAG) and diacylglycerides (DAG), respectively. The significance of the activity of AGK to overall mitochondrial function is that phospholipids, particularly cardiolipins, in the membranes of these organelles are involved in the regulation of mitochondrial apoptosis and autophagy.
In addition to its kinase activity, AGK serves as a component of the translocase of the mitochondrial inner membrane 22 (TIM22) complex and in this complex it facilitates protein anchoring to the mitochondrial inner membrane. This function of AGK does not involve its kinase function. Additional non-kinase activities have been found to be associated with AGK that involve its localization to the plasma membrane and to the cytosol. When localized in the plasma membrane AGK associates with the phosphoinositide phosphatase, PTEN. When AGK is associated with PTEN the phosphatase activity of PTEN is inhibited which allows the PI3K, AKT/PKB, and mTORC1 signal transduction pathways to remain active.
Acylglycerol kinase is encoded by the AGK gene. The AGK gene is located on chromosome 7q34 and is composed of 18 exons that generate two alternatively spliced mRNAs encoding precursor proteins of 422 amino acids (isoform 1) and 350 amino acids (isoform 2). Expression of the AGK gene is ubiquitous with the highest levels of expression found in the intestines, kidney, and brain.
Phospholipases and Phospholipid Remodeling
The fatty acid distribution at the C–1 and C–2 positions of glycerol within phospholipids is continually in flux, owing to phospholipid degradation and the continuous phospholipid remodeling that occurs while these molecules are in membranes. Phospholipid turnover results from the action of numerous different phospholipases and phospholipid phosphatases. There are various phospholipases that exhibit substrate specificities for different positions in phospholipids.
In many cases the acyl group which was initially transferred to glycerol, by the action of the acyl transferases, is not the same acyl group present in the phospholipid when it resides within a membrane. The remodeling of acyl groups in phospholipids is the result of the action of enzymes of the phospholipase A1 (PLA1) and phospholipase A2 (PLA2) families. The products of the PLA1 and PLA2 catalyzed reactions are termed lysophospholipids (LPL or lysoPL).
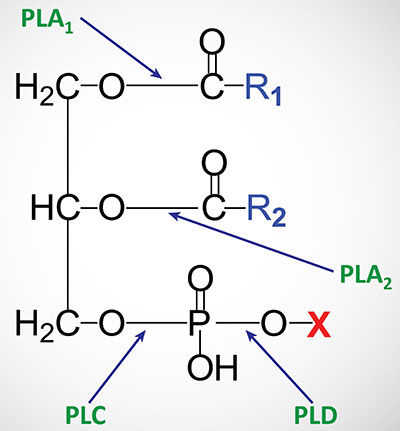
Various lysophospholipids (lysoPL) exhibit biological activities as detailed in the Bioactive Lipids and Lipid Sensing Receptors page as well as the Signal Transduction Pathways: Phospholipids page. In addition to exhibiting biological activities, lysoPL represent an intermediate in the remodeling process of phospholipids. The various lysoPL can be re-esterified by enzymes of the lysophospholipid acyltransferase (LPLAT) family to generate phospholipids with different fatty acid compositions. This pathway of phospholipid synthesis is referred to as the Lands pathway. As discussed above in the Acylglycerophosphate acyltransferases: AGPAT section, humans express 14 genes encoding enzymes of the LPLAT family which includes enzymes also classified as AGPATs and membrane bound O-acyltransferases (MBOATs).
Outside of the context of phospholipid remodeling PLA2 is an important enzyme, whose activity is responsible for the release of polyunsaturated fatty acids (PUFAs) such as arachidonic acid from the C-2 (sn-2) position of membrane phospholipids. The released arachidonate is then a substrate for the synthesis of the eicosanoids. In fact there is not just a single PLA2 enzyme. At least 30 enzymes have been identified with PLA2 activity. For more details on the PLA2 family of lipases visit the Bioactive Lipids and Lipid Sensing Receptors page.
There are 10 isozymes that are in the secretory pathway and these PLA2 isozymes are abbreviated sPLA2. These secretory enzymes are low molecular weight proteins that are Ca2+-dependent and are involved in numerous processes including modification of eicosanoid generation, host defense, and inflammation.
The cytosolic PLA2 family (cPLA2) comprises three isozymes with cPLA2α being an essential component of the initiation of arachidonic acid metabolism. Like the sPLA2 enzymes, the cPLA2 enzymes are tightly regulated by Ca2+. In addition, this class of PLA2 enzyme is regulated by phosphorylation. An additional family of two PLA2 isozymes that are Ca2+-independent for activity are identified as iPLA2. This latter class of enzyme is involved primarily with the remodeling of phospholipids.
Finally, a class of PLA2 enzymes, whose original member was identified as being involved in the hydrolysis and inactivation of platelet activating factor, PAF (see the section below), contains at least four members. The original activity was called PAF-acetylhydrolase (PAF-AH). The PAF hydrolyzing PLA2 isozymes are Ca2+-independent like the iPLA2 family. Because this latter family was shown to not only hydrolyze PAF but also oxidized phospholipids and to be associated with lipoprotein particles in the circulation they are identified as the lipoprotein-associated PLA2 (Lp-PLA2) family. More details on the functions of the Lp-PLA2 family of enzymes can be found in the Lipoproteins, Blood Lipids, and Lipoprotein Metabolism page.
Plasmalogens
Plasmalogens are glycerol ether phospholipids. They are of two types, alkyl ether (–O–CH2–) and alkenyl ether (–O–CH=CH–). Dihydroxyacetone phosphate serves as the glycerol precursor for the synthesis of glycerol ether phospholipids. Three major classes of plasmalogens have been identified: choline, ethanolamine and serine plasmalogens. Ethanolamine plasmalogen is prevalent in myelin. Choline plasmalogen is abundant in cardiac tissue.
The initiation of plasmalogen synthesis takes place inside the lumen of the peroxisome using dihydroxyacetone phosphate (DHAP) as the building block. The first enzyme in the synthesis of plasmalogens is glycerone phosphate O-acyltransferase encoded by the GNPAT gene (also known as dihydroxyacetone phosphate acyltransferase, DHAPAT). The GNPAT enzyme adds a fatty acyl group to the sn-1 position of DHAP.
The GNPAT gene is located on chromosome 1q42.2 and is composed of 16 exons that generate two alternatively spliced mRNAs encoding proteins of 680 amino acids (isoform 1) and 619 amino acids (isoform 2).
The next step in the synthesis pathway is catalyzed by alkylglycerone phosphate synthase encoded by the AGPS gene. The AGPS encoded enzyme exchanges the acyl group added by GNPAT for an alkyl group. The alky-DHAP is then reduced by a reductase found in both the peroxisomes and the ER.
The AGPS gene is located on chromosome 2q31.2 and is composed of 22 exons that encode a 658 amino acid precursor protein.
The remaining reactions of plasmalogen synthesis occur within the ER and include acylation reactions at the sn-2 position and the removal of the phosphate group by one of the phosphatidic acid phosphatase family of enzymes.
One particular type of choline plasmalogen (1-O-1′-enyl-2-acetyl-sn-glycero-3-phosphocholine) has been identified as an extremely powerful biological mediator, capable of inducing cellular responses at concentrations as low as 10–11M. This molecule is called platelet activating factor, PAF. PAF functions as a mediator of hypersensitivity, acute inflammatory reactions and anaphylactic shock. PAF is synthesized in response to the formation of antigen-IgE complexes on the surfaces of basophils, neutrophils, eosinophils, macrophages and monocytes. The synthesis and release of PAF from cells leads to platelet aggregation and the release of serotonin from platelets. PAF also produces responses in liver, heart, smooth muscle, and uterine and lung tissues.
Clinical significance of the plasmalogen biosynthesis pathway is evidenced by the severe phenotypes associated with deficiencies in enzymes of the pathway. Deficiencies in both GNPAT and AGPS result in the peroxisomal disorders of the rhizomelic chondrodysplasia punctata (RCDP) family. These disorders are identified as RCDP1, RCDP2, RCDP3, RCDP4, and RCDP5.
The various RCDP disorders are characterized by disproportionately short stature primarily affecting the proximal parts of the extremities, a typical facial appearance including a broad nasal bridge, epicanthus, high-arched palate, dysplastic external ears, and micrognathia, congenital contractures, characteristic ocular involvement, dwarfism, and severe intellectual impairment with spasticity.
Almost all RCDP infants die within the first year of life. The most commonly occurring RCDP is RCDP1. Whereas RCDP1 and RCDP5 are classified as peroxisomal biogenesis disorders, RCDP2, RCDP3, and RCDP4 are classified as single peroxisomal enzyme deficiencies. Mutations in the PEX7 gene are the causes of RCDP1 and mutation in the PEX5 gene are the causes of RCDP5. Mutations in the GNPAT gene result in RCDP2, mutations in the AGPS gene result in RCDP3, and mutations in the FAR1 gene result in RCDP4.

